
Genetics and biology of pancreatic cancer and its precursor lesions: lessons learned from human pathology and mouse models
Pancreatic ductal adenocarcinoma (PDA) has one of the worst prognoses of human cancers and it is the fourth leading cause of cancer-related deaths in both the US and Japan (1,2). Worldwide, PDA accounts for more than 200,000 deaths per year, with the total number of PDA-related deaths currently increasing and PDA predicted to be the second leading cause of cancer-related deaths in the US by 2030 (3). To improve its prognosis, new diagnostic and treatment strategies are urgently required; thus, a better understanding of the genetics and biology of PDA and its precursor lesions is important. Here, we review the current knowledge regarding the genetics and biology of PDA and its precursor lesions, mainly obtained through mouse model studies.
Genomic events in PDA
Recent genomic sequencing analyses have revealed the mutational landscape of PDA, which has four common oncogenic events in well-known cancer genes (KRAS, TP53, SMAD4, and CDKN2A; known as the “Big 4”) (4-8). In particular, gain-of-function KRAS gene mutations occur in most PDA cases (>90%). Mutations in genes not included in the “Big 4” occur with a relatively low prevalence. Significantly mutated genes can be grouped into several core signaling pathways. The initial whole exome sequencing study of 24 PDA samples identified 12 core signaling pathways involved in PDA: KRAS signaling, TGF-β signaling (including SMAD4), JNK signaling, integrin signaling, WNT/NOTCH signaling, Hedgehog signaling, control of G1/S phase transition (including CDKN2A and TP53), apoptosis, DNA damage control, small GTPase-signaling, invasion, and homophilic cell adhesion (4). The second whole exome sequencing study of 99 PDA samples identified axon guidance, including SLIT/ROBO signaling, as a novel pathway involved in PDA (5). Resent whole genome sequencing data on 456 PDA samples grouped the mutated genes into ten molecular mechanisms: activating KRAS mutations, disruption of G1/S checkpoint machinery, TGF-β signaling, NOTCH signaling, WNT signaling, chromatin modification, the SWI/SNF complex, DNA-damage repair genes, SLIT/ROBO signaling, and RNA processing genes (8). These findings imply that these pathways are important for PDA development and/or progression, although their functions need to be explored further.
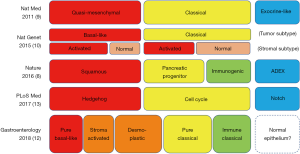
Recently, the transcriptomic subtyping of PDA has also been performed. The initial study defined three PDA subtypes: classical, quasi-mesenchymal (QM), and exocrine-like (9). The QM-PDA subtype has a high tumor grade and a worse prognosis, whereas the classical subtype has high GATA6 expression and is KRAS-dependent. Then, computationally microdissecting approach defined two PDA tumor subtypes: basal-like and classical, and two stromal subtypes: normal and activated (10). The basal-like subtype and activated stroma in the classical subtype are associated with poor survival. Moreover, another large-scale analysis of transcriptomic data of 266 PDA samples defined four PDA subtypes: squamous, immunogenic, pancreatic progenitor, and aberrantly differentiated endocrine exocrine (ADEX) (8). The squamous subtype features ΔNp63, an isoform of p63, and is associated with mutations in chromatin modification genes such as MLL2, MLL3, and KDM6A. This subtype also loses endodermal identity via the methylation of endodermal genes, including HNF4A and GATA6. The squamous subtype has worse prognosis and correlates well with the QM-PDA and basal-like subtypes described above (8,11). The immunogenic subtype is correlated with low tumor cellularity, more pronounced immune response gene expression, and is associated with pancreatic progenitor or classical subtypes. The ADEX subtype and exocrine-like subtype described above are also correlated with low tumor cellularity and recent studies suggest that they may be due to the contamination of normal cells (11,12). Composite analysis of these studies redefined three subtypes: Hedgehog (associated with QM-PDA, basal, activated stroma, or squamous), Notch (associated with exocrine-like, normal stroma, or ADEX), and cell cycle (associated with classical or pancreatic progenitor) (13). Furthermore, a recent study using formalin-fixed paraffine embedded samples redefined the PDA subtypes by tumor cellularity, defining three subgroups in high tumor cellularity samples: pure-basal, pure-classical, and immune-classical, and adding two subgroups by analyzing all the samples: desmoplastic and stroma-activated (12). Although the subtyping details differ between these studies, they largely overlap and define the novel biology of PDA (Figure 1). Further details have been well reviewed elsewhere (14).
Genomic instability is another characteristic of cancer. PDA can be classified into four groups on the basis of structural genome variation: stable, scattered, locally rearranged, and unstable. The unstable subtype is associated with DNA damage response (DDR) gene deficiency and therapeutic implications for platinum and poly (ADP-ribose) polymerase (PARP) inhibitors (7). Details are described in later sections of this review.
Precursor lesions of PDA
PDA is generally thought to arise from three representative precursor lesions: pancreatic intraepithelial neoplasia (PanIN), intraductal papillary mucinous neoplasm (IPMN), and mucinous cystic neoplasm (MCN), although MCN is a rare precursor population (15).
PanIN is a microscopic, flat (or papillary), noninvasive epithelial neoplasm characterized by varying mucin levels and varying degrees of cytologic and architectural atypia (16,17). According to a previous classification system, PanIN was classified as PanIN1, PanIN2, and PanIN3 (carcinoma in situ) by the degree of atypia. In a recent expert consensus meeting, the classification was changed to a two-tiered system consisting of low-grade and high-grade PanINs to improve interpretation in a clinical setting (18). PanIN is the most frequent PDA precursor lesion; in autopsy examinations, PanIN-3 was found in 4% of cases, whereas PanIN-1 and PanIN-2 were present in 77% and 28%, respectively (19). Recent genomic analyses have revealed that PanIN-2 and PanIN-3 can spread throughout the entire ductal epithelium by traveling to distinct ductal epithelia (20). Furthermore, PanIN-derived PDA exhibits worse prognosis than IPMN-derived PDA, suggesting that they differ biologically (21-23). However, it is possible that IPMN-derived PDA can be identified earlier during imaging follow-ups.
IPMN is a grossly visible, predominantly papillary (or rarely flat), noninvasive mucin-producing epithelial neoplasm arising in the main pancreatic duct or branch ducts (17). IPMN has also been organized into a three-tiered classification system by the degree of atypia: low-grade, intermediate-grade, and high-grade IPMNs (carcinoma in situ) (17). However, the classification system has recently been changed to a two-tiered system of low-grade and high-grade IPMNs (carcinoma in situ) (18). IPMN has also been classified into four subtypes based on its architecture and mucin expression patterns: gastric, intestinal, pancreatobiliary, and oncocytic (24). IPMN is a risk factor for PDA, since IPMN itself can progress to PDA (IPMN-derived PDA) and conventional PDA can arise at a different site from IPMN (IPMN-concomitant PDA) (25). IPMN-concomitant PDA patients have more microscopic neoplastic lesions (with KRAS mutations) than IPMN-derived PDA patients, which may explain why IPMN is a risk factor for concomitant PDA (26).
Genetic alterations in PanIN
PanIN genomic analysis has revealed that KRAS mutations occur in 90% of PanIN1 lesions; however, the variant allele frequency of KRAS mutations is relatively low in PanIN1 and higher in high-grade PanIN lesions (27). These data suggest that KRAS mutations are the earliest step in PanIN formation and that low-grade PanIN represents heterogeneous lesions including KRAS non-mutant cells. This finding was confirmed using a chimeric mouse model of PanIN (28). Furthermore, recent analysis revealed that KRAS gene amplification occurs even in low-grade PanIN (29). In contrast, alterations in CDKN2A, TP53, and SMAD4 are observed during later stages of human PanIN development (30). These results suggest that PDA develops via the stepwise accumulation of mutations. A recent genetic study provided new insights into this stepwise accumulation, revealing that 65% of PDA cases harbored at least one chromothripsis event, each of which induces thousands of chromosomal rearrangements, and that 16% of PDA cases exhibit a chromothripsis-mediated simultaneous knockout of the “Big 4” genes (31).
In mice, specific KrasG12D allele expression in embryonic pancreatic progenitor cells (PDX1- or PTF1A-expressing cells) led to sporadic PanIN formation that progressed to PDA during long-term observation (32). Moreover, the additional expression of Tp53R172H or Cdkn2a knockout in embryonic pancreatic progenitor cells rapidly progresses to PDA with high prevalence (33,34). These genetically engineered mouse models (GEMMs) mimic human PDA formation well and shed light on PDA development.
Genetic alterations in IPMN
Exome sequencing studies have shown that IPMN has recurrent mutations in KRAS, GNAS, and RNF43 (35,36). The mutational frequencies of KRAS and GNAS differ among subtypes, with meta-analysis revealing that the KRAS mutation frequency was 73% in gastric, 44% in intestinal, 72% in pancreatobiliary, and 29% in oncocytic types. The GNAS mutation frequency was 53% in gastric, 74% in intestinal, 24% in pancreatobiliary, and 15% in oncocytic types (37). In this pooled analysis, the RNF43 mutation frequency was 22.9% and was not associated with the clinicopathologic features of IPMN patients (37).
A recent study investigated the molecular mechanisms of IPMN-derived PDA and IPMN-concomitant PDA (26). The target sequence revealed that TP53 abnormalities are less frequently observed in IPMN-derived PDA than in IPMN-concomitant PDA (33% vs. 71%, respectively) and that GNAS mutations are observed only in IPMN-derived PDA (67%). In IPMN-derived PDA, IPMN subclones with RNF43 and CTNNB1/β-catenin abnormalities were not likely to be selected during tumor progression. Furthermore, IPMN-concomitant PDA can be classified into two subclasses: “de novo” and “branch off”. The “branch off” type has the same KRAS status as coexisting IPMN, shares a common founder clone with coexisting IPMN, and exhibits longer disease-free survival after resection.
In mice, the embryonic pancreatic expression of the GnasR201H mutation with oncogenic Kras resulted in IPMN formation and early death by severe inflammation (38). An inducible GnasR201C model was recently developed that allows IPMN to develop and progress into PDA (39,40). Pancreatic-specific GnasR201C induction in 4-week-old mice with an oncogenic Kras background led to IPMN formation and rapid death, similar to embryonic Gnas activation (39). However, the GnasR201C induction of 8-week-old mice led to PDA development (40). Moreover, sequential GnasR201C induction with adult acinar cell-specific oncogenic Kras and Tp53 heterozygous deletion led to IPMN-derived PDA formation (39). These results suggest that GNAS mutations have an oncogenic role. Moreover, the loss of Smad4, Brg1, Arid1a, Acvr1b, Tff2, or Pten has been reported to induce IPMN formation (41-48).
Cellular origin of PanIN
The cellular origin of PDA is controversial (Table 1). Lineage tracing experiments in adult acinar cell-specific KrasG12D-expressing mice using Ela-CreERT, Mist1CreERT2, or Ptf1aCreER drivers resulted in spontaneous PanIN formation (49,53,55), demonstrating that PanIN is derived from acinar cells. Lineage tracing experiments have also shown that pancreatitis or oncogenic Kras activation can dedifferentiate pancreatic acinar cells into oval tubular complexes, known as acinar-to-ductal metaplasia (ADM) (49-51). In the setting of acute pancreatitis, this dedifferentiation is a transient event and acinar cells are regenerated in wild-type mice; however, under oncogenic Kras expression, dedifferentiation persists and results in PanIN formation (51). Although direct conversion from ADM to PanIN has not been demonstrated, these studies suggest that acinar cells are precursors of PanIN through ADM.
Table 1
| Cellular origin | Genotype | Phenotype | Ref. |
|---|---|---|---|
| Acinar cells | Ela-CreERT; KrasG12D | ADM and PanIN formation | (49-51) |
| Ela-CreERT; KrasG12D; Trp53R172H/+ | PDA formation in the setting of pancreatitis | (52) | |
| Mist1CreERT2; KrasG12D | ADM and PanIN formation | (49,53) | |
| Mist1CreERT2; KrasG12D; Trp53R172H/+ | PDA formation | (54) | |
| Ptf1aCreER; KrasG12D | ADM and PanIN formation | (55) | |
| Ptf1aCreER; KrasG12D; Trp53R172H/+ | PanIN and PDA formation | (56) | |
| Ptf1aCreER; KrasG12D; Trp53flox/flox | PDA formation | (57) | |
| Ptf1aCreER; KrasG12D; Trp53loxP/+; TetO-GNASR201C | IPMN-derived PDA formation | (39) | |
| Ductal cells | Sox9-CreER; KrasG12D | A few PanIN formation. 112-fold less than Ptf1aCreER; KrasG12D mice | (55) |
| Sox9-CreER; KrasG12D; Trp53flox/flox | Rapid PDA formation and few high grade PanIN formation | (57) | |
| CK19CreERT; KrasG12D | A few PanIN formation | (58) | |
| Ck19CreERT; KrasG12D; Trp53flox/flox | Ductal atypia and rapid PDA formation | (28) | |
| Hnf1b-CreERT2; KrasG12D; Trp53R172H/+ | Normal | (54) | |
| Hnf1b-CreERT2; KrasG12D; Trp53R172H/R172H | Rapid PDA formation without PanIN formation | (54) | |
| Hnf1b-CreERT2; KrasG12D; Brg1flox/flox | IPMN formation | (42) | |
| Hnf1b-CreERT2; KrasG12D; Arid1aflox/flox | IPMN formation | (43) | |
| Sox9-CreER; KrasG12D; Arid1aflox/flox | IPMN and PDA formation | (45) | |
| Sox9-CreER; Ptenflox/flox | IPMN and IPMN-derived PDA formation | (48) | |
| Sox9-CreER; KrasG12D; Ptenflox/+ | PanIN, Pre-IPMN, IPMN and PDA formation | (48) | |
| Sox9-CreER; KrasG12D; Ptenflox/flox | Rapid IPMN and IPMN-derived PDA formation. Died within 2–4 weeks | (48) | |
| Insulin producing cells (extra islet) | RIP-CreERTM; KrasG12D | PanIN formation in the setting of pancreatitis | (59) |
| RIP-CreERTM; KrasG12D; Trp53flox/flox | Undifferentiated PDA formation in the setting of pancreatitis | (59) |
PDA, pancreatic ductal adenocarcinoma; ADM, acinar-to-ductal metaplasia; PanIN, pancreatic intraepithelial neoplasia; IPMN, intraductal papillary mucinous neoplasm.
Other lineage tracing studies in adult ductal cell-specific KrasG12D expressing-mice using CK19CreERT (marker of large ductal cells) and Sox9-CreER (marker of ductal and centroacinar cells) drivers showed that ductal cells can give rise to PanIN, albeit at an extremely low frequency (55,58). The latter study found that PanIN from ductal cells is located near large ducts and occurs at a frequency of less than 100-fold compared with PanIN from acinar cells (55). Recent studies have shown that PDA without concomitant IPMN lesions can be developed from ductal cells if there are deletions or mutations in TP53 or deletion of Fbxw7 alleles with KrasG12D expression. They concluded that duct-derived PDA was more lethal than acinar-derived PDA (28,54,57); however, early PanIN lesions were not observed in these mouse models, suggesting that PDA of ductal origin has a distinct progression mechanism from the traditional PanIN model.
Another lineage tracing study in insulin-producing cell-specific KrasG12D expressing-mice using a RIP-CreERTM driver showed that PanIN can originate from extra-islet insulin-producing cells, not spontaneously, but under pancreatic inflammation (59).
Cellular origin of IPMN
IPMN is a neoplasm arising in the main pancreatic duct or branch ducts of humans (17). Several mouse models of IPMN have been developed using duct-specific Cre lines and show that mouse IPMN is derived from ductal cells (42,43,45,48). Another study suggested that IPMN can be derived from pancreatic duct glands (PDG), which are gland-like outpouches budding off the main pancreatic ducts that function as a progenitor niche for the ductal epithelium and express TFF2 (60,61). Embryonic global Tff2 knockout with pancreatic-specific KrasG12D expression led to PDG hyperplasia and the formation of IPMN-like lesions which progress to PanIN lesions via Smad4 regulation (47). However, a recent study showed that acinar cell-specific Gnas mutations can lead to IPMN (39), suggesting that GNAS mutations result in IPMN regardless of the cellular origin.
Cellular origin of human PDA
Evidence suggests that PanIN is mostly derived from acinar cells; however, PDA can also originate from ductal cells and insulin-producing cells in mice. Recent studies using an organoid culture system have provided further insight into the cellular origin of human PanIN and PDA. CRISPR/Cas9 enabled oncogenic KRAS overexpression and the deletion of CDKN2A, TP53, and SMAD4 to be engineered in human duct cell organoids, which subsequently developed PanIN-like lesions after orthotopic transplantation (62). Another group performed oncogenic KRAS knock-in and the deletion of CDKN2A, TP53, and SMAD4 in pancreatic ductal organoids, which developed PDA after subcutaneous transplantation (63). These data suggest that human ductal cells can give rise to PDA, however, the contribution of human acinar cells towards PDA development remains unclear.
Inflammation and PanIN/PDA
Chronic pancreatitis is a risk factor for PDA (64). Experimental pancreatitis induced by caerulein, a cholecystokinin analog, caused ADM and accelerated PanIN formation in an embryonic KrasG12D expression model. Chronic pancreatitis in an adult acinar cell-specific KrasG12V expressing model with CDKN2A or TP53 deletions induced high-grade PanIN and invasive PDA by overcoming oncogene-induced senescence, even though these mice only spontaneously induce low-grade PanIN formation (65). This study shows that inflammation is a critical step in PanIN/PDA progression, a notion supported by many studies in which the knockout of key inflammatory regulators, including Nfκb, IL-6, and Stat3, induced the loss of PanIN formation in a mutant Kras background (66-69). Furthermore, chronic inflammation induced by COX2 or IKK2 activation in a TP53-null background produced various pancreatic cancer subtypes, including acinar cell carcinoma, PDA, sarcomatoid carcinoma, and neuroendocrine tumors without mutant Kras (70). These findings suggest that chronic inflammation plays an important role in PDA formation independently of Kras mutations.
Many studies have been conducted to understand the roles of environmental factors, including stromal cells, hematopoietic cells, and immune cells, in PDA formation. Tumor-stroma and tumor-immune cell interactions are hot topics in pancreatic cancer research and have been reviewed in detail elsewhere (71,72).
Involvement of molecular signaling pathways in PDA development
Resent whole genome sequencing data on 456 PDA samples showed that significantly mutated genes in human PDA can be grouped into ten molecular mechanisms (8). Here, we describe the current understanding of these signaling pathways in PDA and their precursor lesions (Table 2).
Table 2
| Pathway | Gene | Initiation (Solely) | Initiation and progression (with Kras mutation) | Progression of lesions | Ref. |
|---|---|---|---|---|---|
| KRAS and RTK signaling | KRAS | Oncogenic | – | Oncogenic (in PDA) | (32,73-76) |
| BRAF | Oncogenic | – | – | (77) | |
| CRAF | – | Oncogenic | – | (78) | |
| PIK3CA | Oncogenic | Oncogenic | – | (78,79) | |
| PDK1 | – | Oncogenic | Oncogenic (in PanIN) | (78,80) | |
| RAC1 | – | Oncogenic | – | (79) | |
| TRAIL-R | – | Oncogenic | – | (81) | |
| PTEN | Tumor suppressive (IPMN) | Tumor suppressive | – | (48,82-84) | |
| RICTOR | – | Oncogenic | – | (85) | |
| EGFR | – | Oncogenic | – | (86,87) | |
| PTPN11 (SHP2) | – | Oncogenic | Oncogenic (in PDA) | (88) | |
| CRAF + EGFR | – | Oncogenic | Oncogenic (in PDA) | (89) | |
| Cell cycle | CDKN2A | – | Tumor suppressive | – | (34) |
| TP53 | Tumor suppressive (under inflammation) | Tumor suppressive | – | (33,65) | |
| RB | – | Tumor suppressive | – | (90) | |
| p21 (CDKN1A) | – | Tumor suppressive | – | (91) | |
| MYC | Oncogenic | Oncogenic | – | (92-97) | |
| TGF-β signaling | SMAD4 | – | Context dependent1) | – | (41,98-100) |
| TGFBR2 | – | Tumor suppressive | – | (101) | |
| ACVR1B | – | Tumor suppressive (IPMN) | – | (102) | |
| NOTCH signaling | NOTCH1 | – | Context dependent2) | – | (50,96,103) |
| NUMB | – | Tumor suppressive | – | (104) | |
| NOTCH2 | – | Oncogenic | – | (96) | |
| LFNG | – | Tumor suppressive | – | (105) | |
| DNMAML | – | Tumor suppressive | – | (106) | |
| SOX9 | – | Oncogenic | – | (55) | |
| HES1 | – | Context dependent3) | – | (52,107) | |
| FBXW7 | – | Tumor suppressive | – | (28,108) | |
| WNT signaling | β-catenin | Oncogenic (SPN) | Context dependent4) | – | (51,109-111) |
| DKK1 | – | Oncogenic | Oncogenic (in PanIN) | (110) | |
| ATDC | – | Oncogenic | – | (112) | |
| WNT1 | – | Oncogenic (PanIN, MCN) | – | (111) | |
| NFATc1 | – | Oncogenic | – | (113-115) | |
| Chromatin modification | KDM6A | – | Tumor suppressive | – | (116) |
| BMI1 | – | Oncogenic | – | (117) | |
| RING1B | – | Oncogenic | Oncogenic (in PDA) | (118) | |
| EZH2 | – | Tumor suppressive | – | (119) | |
| SWI/SNF complex | SMARCA4 (Brg1) | – | Context dependent5) | Oncogenic (in PanIN) | (42,56,120) |
| ARID1A | – | Tumor suppressive | – | (43-45,121) | |
| SMARCB1 | – | Tumor suppressive | Tumor suppressive (in PDA) | (122) | |
| DNA-damage repair genes | BRCA1 | – | Tumor suppressive | – | (123,124) |
| BRCA2 | – | Context dependent6) | – | (125-129) | |
| ATM | – | Tumor suppressive | – | (130,131) | |
| SLIT/ROBO signaling | ROBO2 | – | No effect | – | (132) |
1), mainly a tumor suppressive role but heterozygous mutation with TP53 mutation inhibits metastasis; 2), knockout models show a tumor suppressive role, whereas overexpression model shows an oncogenic role; 3), embryonic pancreatic knockout model shows a tumor suppressive role, but adult acinar specific knockout model shows an oncogenic role; 4), mainly an oncogenic role but simultaneous β-catenin stabilization with Kras mutation leads ITT formation and impairs PanIN formation; 5), a tumor suppressive role in ductal cell-derived IPMN formation but an oncogenic role in acinar cell-derived PanIN formation. PDA, pancreatic ductal adenocarcinoma; IPMN, intraductal papillary mucinous neoplasm; PanIN, pancreatic intraepithelial neoplasia; MCN, mucinous cystic neoplasm; ITT, intraductal tubular tumor.
KRAS and RTK signaling
KRAS mutations are the earliest step in PanIN formation in terms of the mutational landscape of PDA, as described previously (27). KRAS is a key player in the RTK/RAS signaling pathway and its mutation causes constitutive Ras-GTP activation leading to downstream signaling.
In inducible KrasG12D mouse models which allow reversible and conditional KrasG12D expression, Kras mutations were shown to be indispensable for PanIN initiation and PanIN and PDA progression by controlling anabolic glucose metabolism (73,74). Although the majority of PDA cells undergo apoptosis upon Kras repression, a few PDA cells with stem cell properties can survive via oxidative phosphorylation (75) and in a Yap-dependent manner (76).
One important signal downstream of KRAS is the RAF-MEK-ERK pathway. RAF kinases, including A-RAF, B-RAF, and C-RAF, are direct KRAS effectors that mediate MAPK activation. Activated RAF phosphorylates MEK1 and MEK2, which in turn activate the serine-threonine kinases ERK1 and ERK2. BRAF mutations are found in approximately 3% of human PDA cases and are mutually exclusive with KRAS mutations (6). The pancreas-specific expression of a BrafV600E mutant allele initiated PanIN formation without a Kras mutation and resulted in lethal PDA with a Tp53R270H mutation in mice (77). The effect of the genetic inactivation of the RAF-MEK-ERK pathway in KrasG12V-mediated lung tumors was investigated. The deletion of MEK or ERK by combining Mek1lox/lox and Mek2−/− or Erk1−/− and Erk2lox/lox significantly improved survival; however, systemic elimination in adult tissues led to early mouse mortality (133). Interestingly, whereas Kras-driven lung tumor formation is blocked in the absence of C-Raf in mice without lethality by systemic elimination in adult tissues (133,134), pancreas-specific C-Raf ablation had no suppressive effects on PDA development in mice (78). However, additional Egfr ablation critically inhibited PDA formation and led to the complete regression of 6/12 established PDA tumors in mice without lethal effects (89).
Another important signal downstream of KRAS is the PI3K-PDK-AKT pathway. Embryonic pancreatic or adult acinar cell-specific expression of the Pik3caH1047R mutant allele induced PanIN formation and PDA progression (78). Furthermore, the embryonic pancreatic deletion of Pik3ca or the central downstream factor Pdk1 abolished KrasG12D-mediated PanIN formation in mice (78,79). A recently developed dual recombinase system, which combines flippase-FRT (Flp-FRT) and Cre-loxP recombination technologies to sequentially manipulate gene expression, revealed that PDK1 is also indispensable for PanIN progression (80). The embryonic pancreatic deletion of Pik3ca with oncogenic Kras led to AKT phosphorylation but RAC1 downregulation, which is another factor downstream of PIK3CA. Embryonic pancreatic Rac1 deletion abolished PanIN formation, indicating that RAC1 is a critical factor downstream of PIK3CA in PanIN formation (79). Moreover, the loss of mTRAIL-R, an ortholog of the human TRAIL-receptor, delayed PDA growth, inhibited metastasis, and prolonged the survival of PDA-bearing mice in KrasG12D and Trp53+/− backgrounds via direct RAC1 inhibition (81). The tumor suppressor gene PTEN is a negative regulator of the PI3K pathway. The embryonic pancreatic loss of Pten led to the expansion of centroacinar cells and papillary carcinoma (82), whereas adult acinar cell-specific Pten deletion does not lead to PanIN formation, but adult duct cell-specific Pten deletion does lead to IPMN formation (48). Furthermore, the embryonic pancreatic heterozygous loss of Pten with Kras mutations accelerated PanIN formation and PDA progression by activating downstream mTOR signaling via the mTORC1 and mTORC2 complexes (83,84). The embryonic inactivation of mTORC2 by Rictor deletion, which is an essential subunit of mTORC2, profoundly prolongs the survival of PDA-bearing mice in KrasG12D and Trp53+/− backgrounds (85). Serotonin (5-HT) was recently identified as an upstream regulator of the PI3K/AKT/mTOR pathway; its stimulation promotes HTR2B (a 5-HT receptor that has a selective antagonistic drug)-LYN-p85 complex formation, and activates the PI3K/AKT/mTOR pathway (135).
Signals upstream of Kras are also important for Kras-mediated pancreatic tumorigenesis. Loss of Egfr or the EGFR ligand sheddase Adam17 in the embryonic pancreas blocked the formation of ADM and PanIN with Kras mutations and reduced the formation of PDA with additional Trp53 deletions (86,87). The loss of Ptpn11 (also known as SHP2) significantly inhibited the formation of PanIN with Kras mutations and inhibited PDA formation and progression with additional Trp53 deletions (88). These findings suggest that signals upstream of KRAS are also required to initiate pancreatic tumorigenesis. Consistently, other studies have shown that pancreatic tumorigenesis is strongly dependent on a minimal threshold of KRAS activity which is not achieved simply by a single mutant Kras allele, but by a positive feedback loop with inflammatory stimuli (67) and by amplification (29).
Cell cycle
CDKN2A, TP53, and TP53BP2 are involved in the cell cycle; the roles of CDKN2A and TP53 in pancreatic tumorigenesis are described above. CDKs and RB signaling are also key regulators of the cell cycle, with the embryonic pancreatic loss of Rb with oncogenic Kras shown to accelerate PanIN formation, increase the frequency of cystic neoplasms, and promote rapid PDA formation by inhibiting the senescence response and reducing Tp53 activity (90). P21, also known as CDKN1A, is a CDK inhibitor (136). The embryonic pancreatic heterozygous loss of p21 with oncogenic Kras was shown to accelerate PanIN and PDA formation by overcoming oncogene-induced senescence (91).
MYC stimulates the cell cycle via several parallel mechanisms (92). Myc overexpression in the embryonic acinar compartment under control of the Elastase promoter (Ela-MYC model) resulted in the development of acinar cell carcinomas or mixed carcinoma with acinar and ductal differentiation (93). Embryonic pancreatic MYC activation in an inducible Myc overexpression model led to the development of PDA with liver metastasis, whereas Myc downregulation in established PDA led to cell death and lesion regression (94). However, a recent Myc knock-in model showed that embryonic pancreatic Myc expression does not form tumors by itself, although it significantly reduced survival in an oncogenic Kras background (95). Embryonic pancreatic Myc deletion with oncogenic Kras resulted only in PanIN1 formation (96), whereas embryonic pancreatic Myc heterozygous deletion with oncogenic Kras and TP53 mutation reduced PDA progression and increased survival (95). An ESC-based mouse model using an inducible RNA interference-based approach showed that Myc knockdown in adult pancreatic epithelium in an oncogenic Kras background decreased, but did not abolish, ADM and PanIN formation (97). Mechanistically, MYC regulates ductal-neuroendocrine lineage plasticity in PDA and induces gemcitabine resistance (95). A subset of PanIN cells expressing neuroendocrine markers have been shown to promote PanIN progression in connection with sensory neurons, which are known to cause PanIN and PDA progression (137,138).
TGF-β signaling (including SMAD4)
TGF-β signaling is known to have both pro-tumorigenic and tumor suppressive roles in a context-dependent manner. SMAD4, the central mediator of TGF-β, is inactivated in approximately 55% of human PDA cases (139), whereas other TGF-β signaling members are mutated in less than 10%. Furthermore, SMAD4 inactivation is highly correlated with TP53 inactivation (140).
In mice, the loss of Smad4 in the embryonic pancreas with Kras mutations led to IPMN or MCN formation and PDA progression (41,98,99). Moreover, in the context of Kras and Tp53 mutations, the heterozygous loss of Smad4 in the embryonic pancreas inhibited metastasis compared to Smad4 wild-type mice, whereas the homozygous loss of Smad4 exhibited a similar metastatic burden as in Smad4 wild-type mice, although proliferation was inversely correlated with Smad4 gene dosage (100). When looking at other members of TGF-β signaling, loss of Tgfbr2 (the TGF-β receptor) in the embryonic pancreas with Kras mutations led to highly aggressive (with dense stroma but less metastatic) PDA formation with a short latency time (101). Furthermore, the inhibition of CXCR2 disrupted tumor-stromal interactions and prolonged survival in this mouse model (141). Loss of Acvr1b (activin receptor and another ligand of the TGF-β superfamily) in the embryonic pancreas with Kras mutations led to IPMN formation (46). Additionally, the pharmacological inactivation of TGF-β accelerated PDA initiation and progression in mice (102). These studies demonstrate the tumor suppressive role of TGF-β during the early stages of PDA development. Conversely, TGF-β is known to induce epithelial-mesenchymal transition (EMT), which promotes invasion and metastasis (142). TGF-β also promotes cancer stem cell heterogeneity and drug resistance in squamous cell carcinoma (143). These two papers highlight the context-dependent pro-tumorigenic and tumor suppressive roles of TGF-β signaling in cancer initiation and progression. A recent study revealed that SMAD4 is a critical regulator of this context-dependency in PDA. TGF-β treatment in Smad4-normal PDA promoted apoptosis via SMAD2/3-mediated SOX4 expression and SMAD4-mediated KLF5 repression through SNAIL-driven EMT, whereas TGF-β treatment in Smad4-mutated PDA promoted tumorigenesis via SOX4 and KLF5 cooperation (144).
NOTCH signaling
The NOTCH signaling pathway is a highly evolutionarily conserved pathway that mediates cell-to-cell communication (145) and plays a critical role in cell proliferation, differentiation, development, and homeostasis (146). NOTCH signaling (including JAG1, NF2, BCORL1, and FBXW7) is frequently mutated (8) and multiple components are upregulated in human PDA (147). NOTCH signaling has four receptors: NOTCH1-4. NOTCH1 is expressed in acinar cells, whereas NOTCH2 is expressed in ductal and centroacinar cells (96). Embryonic pancreatic Notch1 deletion with oncogenic Kras accelerated PanIN formation and progression and reduced survival via β-catenin activation (96,103). These results suggest that NOTCH1 has a tumor suppressive role in PDA formation. Interestingly, embryonic pancreatic or adult acinar cell-specific Notch1 activation with oncogenic Kras accelerated ADM and PanIN formation/progression (50). Consistently, the embryonic pancreatic and adult acinar-specific deletion of Numb [a multifunctional protein that negatively regulates NOTCH signaling, especially NOTCH1 (104)] with KrasG12D also accelerated ADM and PanIN formation although it reduced cell viability during PanIN progression (148). These data suggest that the tumor suppressive role of NOTCH1 is dose-dependent. Furthermore, adult acinar cell-specific Notch1 deletion increased the number of PanIN lesions without changing their severity, supporting the tumor-suppressive role of NOTCH1 (149).
In contrast to Notch1 deletion, embryonic pancreatic Notch2 deletion with KrasG12D was shown to decrease PanIN progression and prolong survival by downregulating MYC, suggesting that NOTCH2 has an oncogenic role in PDA formation (96). In addition, MCN and late-occurring anaplastic PDA were found in these mice. The role of NOTCH3 in pancreatic tumorigenesis was investigated indirectly; the embryonic pancreatic deletion of Lfng, which regulates the ligand binding of the NOTCH receptor, with KrasG12D accelerated PDA formation by upregulating NOTCH receptors, especially NOTCH3 (105). These studies suggest that each NOTCH receptor has a distinct role in pancreatic tumorigenesis. The role of NOTCH signaling in PanIN development has also been investigated by inducing the dominant negative form of MAML1 (DNMAML), which represses all canonical Notch-mediated transcription in a cell-autonomous manner. Embryonic pancreatic DNMAML expression with KrasG12D delayed PanIN formation, suggesting tumor suppressive role of NOTCH signaling in broad sentence (106). Pharmacologically, the γ-secretase inhibitor (GSI) broadly inhibits NOTCH signaling. Embryonic pancreatic KrasG12D and Tp53+/− expressing mice treated with GSI were resistant to PDA development, supporting an oncogenic role for NOTCH signaling (150). Furthermore, combining GSI and gemcitabine treatments after PDA formation synergistically and significantly improved anti-tumor efficacy and median survival in mice (151); however, due to the strong gastrointestinal side effects of GSI (152,153), a more specific therapy is warranted. SOX9, a master regulator of pancreatic development, is also a factor downstream of NOTCH in pancreatic development. Loss of Sox9 in adult acinar cells with a KrasG12D background prevented PanIN formation (55). Recently, the role of HES1, a critical factor downstream of NOTCH, was also investigated. Embryonic Hes1 deletion, which is involved in the terminal differentiation of adult acinar cells, with KrasG12D promoted ADM and PDA formation, and decreased survival, while prevented high-grade PanIN formation (107). On the other hand, adult acinar cell-specific Hes1 deletion with KrasG12D and/or Tp53R172H significantly inhibited pancreatitis-induced PanIN formation and blocked PanIN progression with downregulating SOX9 (52). Since the loss of HES1 in adult ductal/centroacinar cells led to ductal-to-acinar metaplastic expansion (154,155), it remains unclear whether the immaturation of acinar cells or recombination of ductal cells in embryonic models can explain this phenotypic discrepancy.
FBXW7 is a component of the SKP1-CULLIN1-Fbox E3 ubiquitin ligase complex, which targets multiple well-known oncoproteins, including NOTCH1, by ubiquitination-mediated destruction (156). In the “Sleeping Beauty” transposon-insertional mutagenesis model to cause random mutations, Fbxw7 mutations accelerate KrasG12D-induced PDA formation with a high frequency (24%) (157). The embryonic pancreatic heterozygous deletion of Fbxw7 with KrasG12D accelerated PanIN formation, whereas the homozygous deletion of Fbxw7 with KrasG12D significantly accelerated PDA formation at 2–3 weeks of age via YAP activation (108). Moreover, the adult duct cell- or acinar cell-specific homozygous deletion of Fbxw7 with KrasG12D resulted in PDA formation, with AGR2 defined as a putative marker of duct-derived PDA (28).
WNT signaling
The WNT signaling pathway is an important embryonic signaling pathway that is required for the proliferation, morphogenesis, and differentiation of several organs (158). Mutations in the RNF43 gene, which inhibits Wnt/beta-catenin signaling by ubiquitinating the Frizzled receptor and targeting it to the lysosomal pathway for degradation (159), occur in 5% of PDA cases (8). RNF43 mutations are also associated with IPMN, as described above. Initial functional study of RNF43 inactivating mutations in PDA showed that RNF43-mutated PDA is dependent on WNT signaling and is effectively inhibited by Porcupine inhibitor (160). Recently, genome wide CRISPR/Cas9 screening showed that FZD5 is a druggable target for PDA with RNF43-inactivating mutations (161). The role of WNT signaling in PDA development has been explored. WNT signaling has a canonical pathway, including β-catenin, and a non-canonical pathway (158,162). The initial studies for understanding the role of WNT signaling, especially β-catenin, have been summarized in another review (163). In brief, embryonic pancreatic β-catenin activation led to the formation of pseudo-papillary neoplasm, a rare pancreatic tumor histopathologically characterized by β-catenin activation. Embryonic pancreatic β-catenin activation with KrasG12D inhibited PanIN formation but led to intraductal tubular tumors (109), whereas acinar-specific β-catenin activation with KrasG12D also impaired PanIN formation after acute pancreatitis (51). On the other hand, embryonic pancreatic β-catenin knockout inhibited the regeneration of acinar cells from pancreatitis-induced ADM (51) and, in the context of Kras mutations, the loss of β-catenin inhibited ADM and PanIN formation (110). These results demonstrate the oncogenic role of β-catenin and suggest that precise β-catenin dosage control is critical for ADM and PanIN formation. Further studies confirmed the necessity of WNT signaling in Kras-induced tumorigenesis. The overexpression of Dkk1, an endogenous inhibitor secreted in WNT signaling, using an inducible Dkk1 allele impaired PanIN formation and progression in Kras mutated pancreatic epithelium, furthermore, it inhibited the proliferation of PanIN and induced apoptosis of surrounding stroma in established PanIN (110). Unlike the initial studies on β-catenin dosage impact, recent studies have shown that the overexpression of WNT signaling has a tumor promoting role in the PanIN-PDA sequence. The embryonic pancreatic activation of Atdc (also known as TPRM29) with KrasG12D promoted PanIN progression and PDA formation by inducing EMT via CD44 upregulation after β-catenin upregulation (112). The sequential postnatal activation of Wnt1 or β-catenin in an elastase-tva-based RCAS-TVA system with embryonic Kras activation resulted in PanIN progression and PDAC formation (111). Additionally, female mice with the sequential postnatal activation of Wnt1, but not β-catenin, with embryonic Kras activation developed MCN via paracrine β-catenin activation in stromal cells (113).
The importance of the non-canonical WNT pathway has also been clarified recently. Non-canonical WNT signaling is mediated by intracellular calcium ions and JNK, which leads to NFAT signaling activation (162). The embryonic pancreatic activation of Nfatc1 with KrasG12D promoted PDA formation by controlling gene expression via enhancer-to-promoter communication through NFATc1-STAT3 complex formation. The embryonic deletion of Nfatc1 with KrasG12D inhibited pancreatitis-induced ADM formation, although it did not affect ADM and PanIN formation without pancreatitis (114). The following studies helped to clarify the precise role of NFATc1 in ADM formation. The EGFR-mediated activation of NFATc1 induced ADM development by forming a complex with c-JUN on the Sox9 promoter and activating SOX9 (115). Nfatc1 expression was silenced by EZH2-mediated histone methylation during acinar cell recovery from ADM (164). Moreover, the embryonic pancreatic activation of Nfatc1 with KrasG12D and Tp53R172H resulted in the progression of dedifferentiated PDA, demonstrating that NFATc1 drives EMT reprogramming and maintains PDA in a stem cell-like state via the SOX2-dependent transcription of EMT and stemness factors. NFATc1-SOX2 complex-mediated PDA dedifferentiation and progression is opposed by antithetical p53-miR200c signaling and inactivation of the tumor suppressor pathway is essential for tumor dedifferentiation and dissemination (165).
Chromatin modification
Histone modification enzymes are mutated in 24% of human PDA cases. Mutations have been observed in the KDM6A, SETD2, and ASCOM complex members MLL2 and MLL3 (8). KDM6A, MLL2, and MLL3 exist in the same complex and drive transcriptional activation via H3K4 methylation and H3K27 demethylation (166). KDM6A is a SWI/SNF-interacting partner involved in the demethylation of lysine residues on histones. Transcriptomic analysis revealed that KDM6A mutations or loss occur in 18% of PDA samples and correlate with the squamous subtype, which has the worst prognosis of the four PDA subtypes (7,8). The embryonic pancreatic loss of Kdm6a with oncogenic Kras induced aggressive and metastatic squamous-like PDA in female mice, whereas in male mice the concomitant loss of the Y chromosome-encoded KDM6 family member UTY, which lacks demethylase activity, resulted in a similar phenotype (116).
SETD2 is a methyltransferase known to mediate H3K36 methylation. Its ability to regulate splicing, DNA methylation, chromosome segregation, and DNA-damage repair suggest a tumor suppressive role of SETD2 (167). Furthermore, the loss of SETD2 caused mRNA processing defects in 25% of genes expressed across the genome (168).
Polycomb repressive complexes (PRC) are epigenetic gene silencers involved in the maintenance of a stem cell state and cancer development. PRC1 mediates H2AK119Ub1 and PRC2 mediates H3K27me3 (169). The embryonic pancreatic loss of Bmi1, a PRC1 component, almost completely abrogated PanIN formation in KrasG12D with or without an Ink4a−/− background (117). RING1B, another component of PRC1, is also critical for KrasG12D mediated tumorigenesis from adult acinar cell, whereas the CRISPR/Cas9 mediated knockout of Ring1b in mouse PDA cells reduced tumorigenicity after orthotopic transplantation (118). These data demonstrate the oncogenic function of PRC1. In contrast, EZH2, a PRC2 component, has a tumor suppressive function, since the embryonic pancreatic loss of Ezh2 with KrasG12D accelerated PanIN formation and progression by enhancing COX2-mediated chronic inflammation (119).
SWI/SNF complex
The SWI/SNF chromatin remodeling complex permits DNA-protein contacts to regulate gene expression (170). SWI/SNF complexes contain 12–15 subunits and comprise two main groups, the BRM/BRG1-associated factor (BAF) (SMARCA2/SMARCA4-associated factor in humans) and the Polybromo-associated (PBAF) complexes. Recent whole exome and genome sequences have shown that at least one subunit of the SWI/SNF complexes, including ARID1A, ARID1B, SMARCA4, SMARCA2, PBRM1, and SMARCB1 are mutated in 14% of human PDA cases (5,7,8).
The initial in vivo functional analyses of SWI/SNF complexes in pancreatic carcinogenesis focused on BRG1 (human; SMARCA4), 1 of the 2 catalytic subunits in SWI/SNF complexes. The embryonic pancreatic deletion of Brg1 with KrasG12D led to the formation of cystic neoplasm that highly resembled human pancreatobiliary type IPMN. Under this background, the IPMN lesions progressed to invasive PDA (42). This IPMN-derived PDA model has better prognosis than the classical model of PanIN-derived PDA with KrasG12D and Tp53+/−, mirroring prognostic trends in human PDA patients. Brg1 null IPMN-derived PDA possesses a distinct molecular signature that supports less malignant features than PanIN-derived PDA. Additionally, the adult duct or acinar-specific deletion of Brg1 with KrasG12D revealed that IPMN lesions were derived from ductal cells. Another study showed that BRG1 blocks the initiation of ductal tumorigenesis by inhibiting the dedifferentiation of ductal cells via SOX9 regulation, whereas BRG1 promotes tumorigenesis in IPMN-derived PDA by supporting a mesenchymal-like landscape (120).
In contrast, our recent work showed that the acinar-specific deletion of Brg1 with oncogenic Kras impaired ADM and PanIN formation independently of Tp53R172H and inhibited PanIN-derived PDA formation in the presence of Tp53R172H via the direct downregulation of SOX9. Furthermore, the ablation of Brg1 in established PanINs using a dual recombinase system resulted in their regression (56). These data demonstrate that the BRG1/SOX9 axis is critical for PanIN-derived PDA development, highlighting the cell-specific and context-dependent roles of BRG1 in PDA initiation and progression.
ARID1A is a subunit of the BAF complex and the most frequently mutated gene in multiple human cancers (171), demonstrating that its protein loss and mutations correlated with the poor survival of PDA patients (6). Recently, several in vivo functional analyses have been performed on ARID1A (43-45,121). Our initial report showed that the embryonic pancreatic deletion of Arid1a with KrasG12D led to IPMN formation that progressed to PDA. The adult duct- or acinar-specific deletion of Arid1a with KrasG12D revealed that IPMN caused by Arid1a deletion was also derived from ductal cells. Functionally, Arid1a loss led to the dedifferentiation of ductal cells and pancreatic ductal dilation by suppressing SOX9 expression. These results highly resembled to those of Brg1-deleted mice; however, the incidence of PDA formation in Arid1a-deleted mice was significantly lower than in Brg1-deleted mice, likely since mTOR pathway activation is lower in Arid1a-deleted IPMN than in Brg1-deleted IPMN, and PanIN was formed from adult acinar cell-specific Arid1a-deleted mice (43). These results confirmed the tumor suppressive role of ARID1A in the pancreas. Recent reports from three other groups have provided additional insights. Firstly, the sequential knockdown of Arid1a using an inducible shArid1a model in adult pancreatic epithelium with KrasG12D resulted in rapid and irreversible PanIN formation, but did not increase PDA formation. ARID1A depletion reduced chromatin accessibility at acinar-specific enhancers, limiting the transcriptional output of acinar master transcription factors (121). Secondly, the embryonic pancreatic deletion of Arid1a with KrasG12D and Tp53+/− led to the formation of IPMN and fully invasive, poorly-differentiated adenocarcinomas with increased EMT gene expression and stem cell identity (44). Thirdly, the ductal cell-specific deletion of Arid1a with KrasG12D and Tp53+/− resulted in PDA formation, whereas the acinar-specific heterozygous deletion of Arid1a with KrasG12D and Tp53+/− accelerated PDA formation. This study showed that Arid1a deletion in ductal cells activates MYC and increases protein synthesis (45).
Another SWI/SNF partner, SMARCB1, has a strong tumor-suppressive function. The deletion of Smarcb1 in the embryonic pancreas in oncogenic Kras with or without a p53-null background markedly accelerated tumorigenesis and increased metastatic spread and mesenchymal reprogramming. The loss of Smarcb1 activated MYC, which drives the anabolic switch and adaptive activation of the ER stress-induced survival pathway (122).
Recent reports have shown that several cancers with mutations in SWI/SNF complex genes have synthetic lethal partners (172). Thus, future studies in PDA with alterations in SWI/SNF complex components are required.
DNA-damage repair genes
DNA-damage repair genes are mutated in 17% of PDA cases, including BRCA1, BRCA2, ATM, and PALB2 mutations (5% germline, 12% somatic) (8). Germline mutations in DNA-damage repair genes are important familial pancreatic cancer susceptibility genes (173). Germline and somatic mutations in DNA-damage repair genes are also important since they have homologous repair deficiencies and are potential targets of platinum therapies and PARP inhibitors based on the BRCAness concept (174). PDA is associated with four mutational signatures, of which 20 were identified from mutational analysis in over 7,000 cancers (175) (BRCA signature, aging, DNA mismatch repair deficiency, and APOBEC family). A recent genomic study showed that 14% of PDA cases had BRCA1, BRCA2, or PALB2 mutations, all of which are associated with a BRCA signature or an unstable genome. Potential DNA-damage repair genes are mutated in up to 24% of PDA cases, with PDA defined by a BRCA signature or an unstable genome thought to have potential DNA-damage repair gene mutations (7).
The function of BRCA genes has been well characterized using mouse models. BRCA1 forms various complexes that function in many biological processes (176). Embryonic pancreatic Brca1 deletion with KrasG12D and Tp53−/− significantly reduced tumor latency. The BRCT domain mutation that disrupts BRCA1 function in the DNA-damage response also significantly reduced tumor latency, however loss of E3 ligase activity, which is thought to regulate multiple tumor suppressive pathways, did not reduce tumor latency. This fact indicates that the central role of BRCA1 in pancreatic tumorigenesis is in DNA-repair (123). Moreover, the DNA-demethylating drug 5-aza-2'-deoxycytidine was shown to enhance the survival of mice harboring embryonic pancreatic Brca1-deletion with KrasG12D and Tp53R270H (124).
BRCA2 is a core mediator of homologous repair in the DNA-damage response (176), with embryonic pancreatic Brca2 knockout leading to pancreatic insufficiency (125). Germline heterozygous Brca2 mutations with embryonic pancreatic KrasG12D expression and Brca2 LOH, or embryonic pancreatic Brca2 knockout with KrasG12D led to reduced tumor burden and pancreatic insufficiency (126,127); however, additional Tp53 mutations or knockout in these backgrounds significantly accelerated PDA formation compared to Brca2 wild-type mice (125-127). These studies clearly demonstrate the context-dependent role of BRCA2 in PDA formation. Furthermore, these studies suggest that germline heterozygous BRCA2 mutations are a potential tumor suppressor. Germline heterozygous Brca2 mutations with embryonic pancreatic KrasG12D expression accelerated tumor burden (126,127). Nonetheless, PARP inhibitors including olaparib, cisplatin, and mitomycin C, are efficient in Brca2-null PDA but not in Brca2-heterozygous PDA (126,127). A recent report using embryonic pancreatic Brca2 knockout mice with KrasG12D and Tp53+/− confirmed the tumor suppressive role of BRCA2 and revealed a mechanism whereby reactive nitrogen species-induced DNA lesions caused genomic instability in the absence of Brca2 (128). Another study showed that PD-L1 and IL-6 combination therapy increased the survival of mice harboring embryonic pancreatic Brca2 knockout with KrasG12D and Tp53R270H (129).
ATM is initially characterized by one of the DDR genes (177) and has a broader ability to integrate and direct various signaling cues to maintain cellular homeostasis (178). Embryonic pancreatic heterozygous Atm knockout with KrasG12D enhanced highly stromal infiltrated ADM and PanIN formation via the activation of BMP4 signaling and reduced survival by enhancing EMT and stem cell signaling (130). Atm deletion leads to chromosomal instability, complex structural rearrangements including chromothripsis, and deregulated DNA integrity checkpoints. Thus, Atm deficiency exhibits synthetic lethality with PARP inhibitors and ATR inhibitors (131). PALB2 has also been confirmed as a tumor suppressor in a mouse model of breast cancer (179,180).
SLIT/ROBO signaling
The Roundabout (ROBO) receptors and their secreted SLIT glycoprotein ligands were originally identified as axon guidance molecules that mediate precise axon path finding and neuronal migration during development (181,182). Recent studies have shown that SLIT/ROBO signaling also acts as a tumor suppressor by suppressing WNT signaling activity and downregulating MET signaling activity (183). Aberrations in SLIT2 and/or ROBO1/2 are observed in 23% of human PDA cases (6% mutation and 18% copy number loss) and low ROBO2 expression levels are correlated with poor survival (5). When sequentially comparing normal mouse pancreas, ADM, and PDA, a progressive decrease in Robo2, progressive increase in Robo1, and no change in Robo3 mRNA expression is observed. A recent study showed that the loss of Robo2 in the embryonic pancreas led to stromal activation and immune cell infiltration via the activation of TGF-β signaling after acute pancreatitis; however, under the KrasG12D background, the loss of Robo2 had no effect on PanIN formation, stromal expansion, or immune cell infiltration (132).
RNA processing genes
RNA splicing is an essential process carried out by major and minor spliceosomes to remove noncoding regions (introns) in pre-mRNA before protein translation (184). The RNA splicing genes SF3B1, U2AF1, and RBM10 are mutated in 16% of human PDA cases (8). SF3B1 and U2AF1 are associated with the U2 snRNP of major spliceosomes. In PDA, SF3B1 mutations aggregate as K700E hotspot mutations (8); K700E hotspot mutations in breast cancer are associated with CDH1 mis-splicing (185), whereas U2AF1 mutations aggregate as S34F hotspot mutations; S34F hotspot mutations in lung adenocarcinoma are associated with the mis-splicing of TSH2, a tumor suppressor of the mTOR pathway (185,186). RBM10 is an RNA-binding protein that exclusively represses splicing (187). RBM10 is located on the X chromosome, and its loss-of-function mutations are the cause of the X-linked recessive disorder TARP syndrome, which mainly affects males (188). RBM10 mutations in PDA are associated with longer survival despite the histological features of aggressive disease (6).
Organoids vs. mouse models
So far, we have described the current understanding of PDA revealed by a number of studies using GEMMs which are valuable for understanding PDA biology. The classical GEMMs have a limitation in studies of PDA development, since genes of interest are activated or deleted at the same time as oncogenic Kras induction using a single Cre-based genetic engineering system. The recent development of a dual recombinase system using Flp-FRT enables the role of genes of interest in established PDA to be investigated. Furthermore, the dual recombinase system enables the investigation of tumor-stroma or tumor-immune interactions by manipulating genes of interest in stroma or myeloid cells using their specific Cre line (189,190).
Although GEMMs are faithful models for determining PDA biology, they have several problems. Firstly, engineering multiple alleles increases the time and cost required for the bleeding and maintenance of GEMMs; however, the recently developed CRISPR/Cas9-mediated in vivo genetic manipulation technique may resolve this problem (191). Secondly, GEMMs are mice, not humans. Recent advances in 3D organoid culture models can mimic and maintain mouse and human epithelial organ structures in a dish (192). Organoid cultures are not only useful for normal epithelial tissue research but for also cancer research; organoids from normal pancreatic ductal cells (193) and PDA (194) have been already established and bring various benefits to PDA research. PDA organoids can rapidly generate and be maintained not only from surgical specimens, but also from small amounts of endoscopic ultrasound-fine needle aspiration (EUS-FNA) samples taken during initial human PDA diagnosis (194-196). Genetic and transcriptomic analyses of advanced cancers could be performed using PDA organoids, which were consistent with those from matched primary PDA (196-198). Organoids are a good platform for drug testing prior to the treatment of patients, since their treatment response correlates well with that of the patients (196,198,199). An initial platform for high-throughput drug screening using organoids has also been developed (200). Furthermore, organoids can be used for biological investigation; for example, the genetic manipulation of key driver mutations in normal ductal organoids could mimic PDA development in vitro, as described above (62,63). Co-culture with fibroblasts revealed that cancer-associated fibroblasts have two sub-populations with distinct roles (201,202).
Current problems in PDA therapy resistance and future aspect
PDA has a weak response to current chemotherapeutic agents. Overcoming the therapeutic resistance of PDA is critical for improving the survival of PDA patients. The therapeutic resistance of PDA is attributed to both cell intrinsic and extrinsic factors (203). The PDA transcriptome can predict drug response and clinical outcome, but its mutational status cannot (8-10,196). Intrinsic factors such as aberrant signaling pathways, epigenetics, post transcriptional modifications, altered metabolism, and tumor heterogeneity, and extrinsic factors such as the extracellular matrix, cancer associated fibroblasts, immune cells, inflammation, cancer stem cell niche, hypoxia, and EMT plasticity can all cause transcriptomic changes and induce therapeutic resistance (204).
In this review, we summarized the recent advances in our understanding of PDA genetics and biology, with a particular focus on tumor cell intrinsic factors. We believe that a better understanding of PDA biology and genetics could lead to the future development of novel diagnostic methods and treatments and could improve survival. The development of next generation GEMMs and organoids could improve our understanding of PDA biology and genetics. We believe that further investigations using these novel tools could lead to overcome the therapeutic resistance of this devastating disease.
Acknowledgments
Funding: This work was supported in part by Grants-in-Aid KAKENHI (19H03639), AMED-PRIME, Japan Agency for Medical Research and Development (19cm6010022h0002), and AMED-Project for Cancer Research and Therapeutic Evolution (P-CREATE), Japan Agency for Medical Research and Development (19cm0106142h0002). It was also supported by the Kobayashi Foundation for Cancer Research, the Research Grant of the Princess Takamatsu Cancer Research Fund, the Mitsubishi Foundation, the Takeda Science Foundation, the Uehara Memorial Foundation, the Mochida Foundation, the Kanae Foundation, and the Novartis Foundation.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Min Li) for the series “Science on Pancreatic Cancer” published in Annals of Pancreatic Cancer. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apc.2019.07.02). The series “Science on Pancreatic Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet 2016;388:73-85. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913-21. [Crossref] [PubMed]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801-6. [Crossref] [PubMed]
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399-405. [Crossref] [PubMed]
- Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17:500-3. [Crossref] [PubMed]
- Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168-78. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Electronic address: andrew_aguirre@dfci.harvard.edu;
- Puleo F, Nicolle R, Blum Y, et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology 2018;155:1999-2013.e3. [Crossref] [PubMed]
- Sivakumar S, de Santiago I, Chlon L, et al. Master Regulators of Oncogenic KRAS Response in Pancreatic Cancer: An Integrative Network Biology Analysis. PLoS Med 2017;14:e1002223 [Crossref] [PubMed]
- Collisson EA, Bailey P, Chang DK, et al. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2019;16:207-20. [Crossref] [PubMed]
- Matthaei H, Schulick RD, Hruban RH, et al. Cystic precursors to invasive pancreatic cancer. Nat Rev Gastroenterol Hepatol 2011;8:141-50. [Crossref] [PubMed]
- Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol 2001;25:579-86. [Crossref] [PubMed]
- Hruban RH, Takaori K, Klimstra DS, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol 2004;28:977-87. [Crossref] [PubMed]
- Basturk O, Yang Z, Tang LH, et al. The high-grade (WHO G3) pancreatic neuroendocrine tumor category is morphologically and biologically heterogenous and includes both well differentiated and poorly differentiated neoplasms. Am J Surg Pathol 2015;39:683-90. [Crossref] [PubMed]
- Matsuda Y, Furukawa T, Yachida S, et al. The Prevalence and Clinicopathological Characteristics of High-Grade Pancreatic Intraepithelial Neoplasia: Autopsy Study Evaluating the Entire Pancreatic Parenchyma. Pancreas 2017;46:658-64. [Crossref] [PubMed]
- Makohon-Moore AP, Matsukuma K, Zhang M, et al. Precancerous neoplastic cells can move through the pancreatic ductal system. Nature 2018;561:201-5. [Crossref] [PubMed]
- Matthaei H, Norris AL, Tsiatis AC, et al. Clinicopathological characteristics and molecular analyses of multifocal intraductal papillary mucinous neoplasms of the pancreas. Ann Surg 2012;255:326-33. [Crossref] [PubMed]
- Poultsides GA, Reddy S, Cameron JL, et al. Histopathologic basis for the favorable survival after resection of intraductal papillary mucinous neoplasm-associated invasive adenocarcinoma of the pancreas. Ann Surg 2010;251:470-6. [Crossref] [PubMed]
- Mino-Kenudson M, Fernandez-del Castillo C, Baba Y, et al. Prognosis of invasive intraductal papillary mucinous neoplasm depends on histological and precursor epithelial subtypes. Gut 2011;60:1712-20. [Crossref] [PubMed]
- Furukawa T, Kloppel G, Volkan Adsay N, et al. Classification of types of intraductal papillary-mucinous neoplasm of the pancreas: a consensus study. Virchows Arch 2005;447:794-9. [Crossref] [PubMed]
- Yamaguchi K, Ohuchida J, Ohtsuka T, et al. Intraductal papillary-mucinous tumor of the pancreas concomitant with ductal carcinoma of the pancreas. Pancreatology 2002;2:484-90. [Crossref] [PubMed]
- Omori Y, Ono Y, Tanino M, et al. Pathways of Progression From Intraductal Papillary Mucinous Neoplasm to Pancreatic Ductal Adenocarcinoma Based on Molecular Features. Gastroenterology 2019;156:647-661.e2. [Crossref] [PubMed]
- Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012;142:730-733.e9. [Crossref] [PubMed]
- Ferreira RMM, Sancho R, Messal HA, et al. Duct- and Acinar-Derived Pancreatic Ductal Adenocarcinomas Show Distinct Tumor Progression and Marker Expression. Cell Rep 2017;21:966-78. [Crossref] [PubMed]
- Mueller S, Engleitner T, Maresch R, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018;554:62-8. [Crossref] [PubMed]
- Maitra A, Adsay NV, Argani P, et al. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol 2003;16:902-12. [Crossref] [PubMed]
- Notta F, Chan-Seng-Yue M, Lemire M, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 2016;538:378-82. [Crossref] [PubMed]
- Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003;4:437-50. [Crossref] [PubMed]
- Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7:469-83. [Crossref] [PubMed]
- Aguirre AJ, Bardeesy N, Sinha M, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 2003;17:3112-26. [Crossref] [PubMed]
- Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med 2011;3:92ra66 [Crossref] [PubMed]
- Furukawa T, Kuboki Y, Tanji E, et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep 2011;1:161. [Crossref] [PubMed]
- Lee JH, Kim Y, Choi JW, et al. KRAS, GNAS, and RNF43 mutations in intraductal papillary mucinous neoplasm of the pancreas: a meta-analysis. Springerplus 2016;5:1172. [Crossref] [PubMed]
- Taki K, Ohmuraya M, Tanji E, et al. GNAS(R201H) and Kras(G12D) cooperate to promote murine pancreatic tumorigenesis recapitulating human intraductal papillary mucinous neoplasm. Oncogene 2016;35:2407-12. [Crossref] [PubMed]
- Patra KC, Kato Y, Mizukami Y, et al. Mutant GNAS drives pancreatic tumourigenesis by inducing PKA-mediated SIK suppression and reprogramming lipid metabolism. Nat Cell Biol 2018;20:811-22. [Crossref] [PubMed]
- Ideno N, Yamaguchi H, Ghosh B, et al. GNASR201C Induces Pancreatic Cystic Neoplasms in Mice That Express Activated KRAS by Inhibiting YAP1 Signaling. Gastroenterology 2018;155:1593-1607.e12. [Crossref] [PubMed]
- Bardeesy N, Cheng KH, Berger JH, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev 2006;20:3130-46. [Crossref] [PubMed]
- von Figura G, Fukuda A, Roy N, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 2014;16:255-67. [Crossref] [PubMed]
- Kimura Y, Fukuda A, Ogawa S, et al. ARID1A Maintains Differentiation of Pancreatic Ductal Cells and Inhibits Development of Pancreatic Ductal Adenocarcinoma in Mice. Gastroenterology 2018;155:194-209.e2. [Crossref] [PubMed]
- Wang W, Friedland SC, Guo B, et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut 2019;68:1245-58. [Crossref] [PubMed]
- Wang SC, Nassour I, Xiao S, et al. SWI/SNF component ARID1A restrains pancreatic neoplasia formation. Gut 2019;68:1259-70. [Crossref] [PubMed]
- Qiu W, Tang SM, Lee S, et al. Loss of Activin Receptor Type 1B Accelerates Development of Intraductal Papillary Mucinous Neoplasms in Mice With Activated KRAS. Gastroenterology 2016;150:218-228.e12. [Crossref] [PubMed]
- Yamaguchi J, Mino-Kenudson M, Liss AS, et al. Loss of Trefoil Factor 2 From Pancreatic Duct Glands Promotes Formation of Intraductal Papillary Mucinous Neoplasms in Mice. Gastroenterology 2016;151:1232-1244.e10. [Crossref] [PubMed]
- Kopp JL, Dubois CL, Schaeffer DF, et al. Loss of Pten and Activation of Kras Synergistically Induce Formation of Intraductal Papillary Mucinous Neoplasia From Pancreatic Ductal Cells in Mice. Gastroenterology 2018;154:1509-1523.e5. [Crossref] [PubMed]
- Habbe N, Shi G, Meguid RA, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A 2008;105:18913-8. [Crossref] [PubMed]
- De La O JP, Emerson LL, Goodman JL, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A 2008;105:18907-12. [Crossref] [PubMed]
- Morris JP 4th, Cano DA, Sekine S, et al. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest 2010;120:508-20. [Crossref] [PubMed]
- Nishikawa Y, Kodama Y, Shiokawa M, et al. Hes1 plays an essential role in Kras-driven pancreatic tumorigenesis. Oncogene 2019;38:4283-96. [Crossref] [PubMed]
- Shi G, Zhu L, Sun Y, et al. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology 2009;136:1368-78. [Crossref] [PubMed]
- Bailey JM, Hendley AM, Lafaro KJ, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene 2016;35:4282-8. [Crossref] [PubMed]
- Kopp JL, von Figura G, Mayes E, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012;22:737-50. [Crossref] [PubMed]
- Tsuda M, Fukuda A, Roy N, et al. The BRG1/SOX9 axis is critical for acinar cell-derived pancreatic tumorigenesis. J Clin Invest 2018;128:3475-89. [Crossref] [PubMed]
- Lee AYL, Dubois CL, Sarai K, et al. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut 2018; [Epub ahead of print]. [PubMed]
- Ray KC, Bell KM, Yan J, et al. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS One 2011;6:e16786 [Crossref] [PubMed]
- Gidekel Friedlander SY, Chu GC, Snyder EL, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell 2009;16:379-89. [Crossref] [PubMed]
- Strobel O, Rosow DE, Rakhlin EY, et al. Pancreatic duct glands are distinct ductal compartments that react to chronic injury and mediate Shh-induced metaplasia. Gastroenterology 2010;138:1166-77. [Crossref] [PubMed]
- Yamaguchi J, Liss AS, Sontheimer A, et al. Pancreatic duct glands (PDGs) are a progenitor compartment responsible for pancreatic ductal epithelial repair. Stem Cell Res 2015;15:190-202. [Crossref] [PubMed]
- Lee J, Snyder ER, Liu Y, et al. Reconstituting development of pancreatic intraepithelial neoplasia from primary human pancreas duct cells. Nat Commun 2017;8:14686. [Crossref] [PubMed]
- Seino T, Kawasaki S, Shimokawa M, et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018;22:454-467.e6. [Crossref] [PubMed]
- Lowenfels AB, Maisonneuve P, Cavallini G, et al. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med 1993;328:1433-7. [Crossref] [PubMed]
- Guerra C, Collado M, Navas C, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell 2011;19:728-39. [Crossref] [PubMed]
- Ling J, Kang Y, Zhao R, et al. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell 2012;21:105-20. [Crossref] [PubMed]
- Daniluk J, Liu Y, Deng D, et al. An NF-kappaB pathway-mediated positive feedback loop amplifies Ras activity to pathological levels in mice. J Clin Invest 2012;122:1519-28. [Crossref] [PubMed]
- Lesina M, Kurkowski MU, Ludes K, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 2011;19:456-69. [Crossref] [PubMed]
- Fukuda A, Wang SC, Morris JP 4th, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 2011;19:441-55. [Crossref] [PubMed]
- Swidnicka-Siergiejko AK, Gomez-Chou SB, Cruz-Monserrate Z, et al. Chronic inflammation initiates multiple forms of K-Ras-independent mouse pancreatic cancer in the absence of TP53. Oncogene 2017;36:3149-58. [Crossref] [PubMed]
- Zhang Y, Crawford HC, Pasca di Magliano M. Epithelial-Stromal Interactions in Pancreatic Cancer. Annu Rev Physiol 2019;81:211-33. [Crossref] [PubMed]
- Neesse A, Bauer CA, Ohlund D, et al. Stromal biology and therapy in pancreatic cancer: ready for clinical translation? Gut 2019;68:159-71. [Crossref] [PubMed]
- Collins MA, Bednar F, Zhang Y, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 2012;122:639-53. [Crossref] [PubMed]
- Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012;149:656-70. [Crossref] [PubMed]
- Viale A, Pettazzoni P, Lyssiotis CA, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014;514:628-32. [Crossref] [PubMed]
- Kapoor A, Yao W, Ying H, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 2014;158:185-97. [Crossref] [PubMed]
- Collisson EA, Trejo CL, Silva JM, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov 2012;2:685-93. [Crossref] [PubMed]
- Eser S, Reiff N, Messer M, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013;23:406-20. [Crossref] [PubMed]
- Wu CY, Carpenter ES, Takeuchi KK, et al. PI3K regulation of RAC1 is required for KRAS-induced pancreatic tumorigenesis in mice. Gastroenterology 2014;147:1405-16.e7. [Crossref] [PubMed]
- Schönhuber N, Seidler B, Schuck K, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med 2014;20:1340-7. [Crossref] [PubMed]
- von Karstedt S, Conti A, Nobis M, et al. Cancer cell-autonomous TRAIL-R signaling promotes KRAS-driven cancer progression, invasion, and metastasis. Cancer Cell 2015;27:561-73. [Crossref] [PubMed]
- Stanger BZ, Stiles B, Lauwers GY, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell 2005;8:185-95. [Crossref] [PubMed]
- Hill R, Calvopina JH, Kim C, et al. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res 2010;70:7114-24. [Crossref] [PubMed]
- Ying H, Elpek KG, Vinjamoori A, et al. PTEN is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-kappaB-cytokine network. Cancer Discov 2011;1:158-69. [Crossref] [PubMed]
- Driscoll DR, Karim SA, Sano M, et al. mTORC2 Signaling Drives the Development and Progression of Pancreatic Cancer. Cancer Res 2016;76:6911-23. [Crossref] [PubMed]
- Ardito CM, Gruner BM, Takeuchi KK, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 2012;22:304-17. [Crossref] [PubMed]
- Navas C, Hernandez-Porras I, Schuhmacher AJ, et al. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell 2012;22:318-30. [Crossref] [PubMed]
- Ruess DA, Heynen GJ, Ciecielski KJ, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med 2018;24:954-60. [Crossref] [PubMed]
- Blasco MT, Navas C, Martín-Serrano G, et al. Complete Regression of Advanced Pancreatic Ductal Adenocarcinomas upon Combined Inhibition of EGFR and C-RAF. Cancer Cell 2019;35:573-587.e6. [Crossref] [PubMed]
- Carrière C, Gore AJ, Norris AM, et al. Deletion of Rb accelerates pancreatic carcinogenesis by oncogenic Kras and impairs senescence in premalignant lesions. Gastroenterology 2011;141:1091-101. [Crossref] [PubMed]
- Morton JP, Jamieson NB, Karim SA, et al. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology 2010;139:586-97, 597.e1-6.
- Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta 2015;1849:506-16. [Crossref] [PubMed]
- Sandgren EP, Quaife CJ, Paulovich AG, et al. Pancreatic tumor pathogenesis reflects the causative genetic lesion. Proc Natl Acad Sci U S A 1991;88:93-7. [Crossref] [PubMed]
- Lin WC, Rajbhandari N, Liu C, et al. Dormant cancer cells contribute to residual disease in a model of reversible pancreatic cancer. Cancer Res 2013;73:1821-30. [Crossref] [PubMed]
- Farrell AS, Joly MM, Allen-Petersen BL, et al. MYC regulates ductal-neuroendocrine lineage plasticity in pancreatic ductal adenocarcinoma associated with poor outcome and chemoresistance. Nat Commun 2017;8:1728. [Crossref] [PubMed]
- Mazur PK, Einwachter H, Lee M, et al. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A 2010;107:13438-43. [Crossref] [PubMed]
- Saborowski M, Saborowski A, Morris JP 4th, et al. A modular and flexible ESC-based mouse model of pancreatic cancer. Genes Dev 2014;28:85-97. [Crossref] [PubMed]
- Izeradjene K, Combs C, Best M, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell 2007;11:229-43. [Crossref] [PubMed]
- Kojima K, Vickers SM, Adsay NV, et al. Inactivation of Smad4 accelerates Kras(G12D)-mediated pancreatic neoplasia. Cancer Res 2007;67:8121-30. [Crossref] [PubMed]
- Whittle MC, Izeradjene K, Rani PG, et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell 2015;161:1345-60. [Crossref] [PubMed]
- Ijichi H, Chytil A, Gorska AE, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev 2006;20:3147-60. [Crossref] [PubMed]
- Hezel AF, Deshpande V, Zimmerman SM, et al. TGF-beta and alphavbeta6 integrin act in a common pathway to suppress pancreatic cancer progression. Cancer Res 2012;72:4840-5. [Crossref] [PubMed]
- Hanlon L, Avila JL, Demarest RM, et al. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res 2010;70:4280-6. [Crossref] [PubMed]
- Beres BJ, George R, Lougher EJ, et al. Numb regulates Notch1, but not Notch3, during myogenesis. Mech Dev 2011;128:247-57. [Crossref] [PubMed]
- Zhang S, Chung WC, Xu K. Lunatic Fringe is a potent tumor suppressor in Kras-initiated pancreatic cancer. Oncogene 2016;35:2485-95. [Crossref] [PubMed]
- Thomas MM, Zhang Y, Mathew E, et al. Epithelial Notch signaling is a limiting step for pancreatic carcinogenesis. BMC Cancer 2014;14:862. [Crossref] [PubMed]
- Hidalgo-Sastre A, Brodylo RL, Lubeseder-Martellato C, et al. Hes1 Controls Exocrine Cell Plasticity and Restricts Development of Pancreatic Ductal Adenocarcinoma in a Mouse Model. Am J Pathol 2016;186:2934-44. [Crossref] [PubMed]
- Zhang Q, Zhang Y, Parsels JD, et al. Fbxw7 Deletion Accelerates Kras(G12D)-Driven Pancreatic Tumorigenesis via Yap Accumulation. Neoplasia 2016;18:666-73. [Crossref] [PubMed]
- Heiser PW, Cano DA, Landsman L, et al. Stabilization of beta-catenin induces pancreas tumor formation. Gastroenterology 2008;135:1288-300. [Crossref] [PubMed]
- Zhang Y, Morris JP 4th, Yan W, et al. Canonical wnt signaling is required for pancreatic carcinogenesis. Cancer Res 2013;73:4909-22. [Crossref] [PubMed]
- Sano M, Driscoll DR, DeJesus-Monge WE, et al. Activation of WNT/beta-Catenin Signaling Enhances Pancreatic Cancer Development and the Malignant Potential Via Up-regulation of Cyr61. Neoplasia 2016;18:785-94. [Crossref] [PubMed]
- Wang L, Yang H, Abel EV, et al. ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev 2015;29:171-83. [Crossref] [PubMed]
- Sano M, Driscoll DR, De Jesus-Monge WE, et al. Activated wnt signaling in stroma contributes to development of pancreatic mucinous cystic neoplasms. Gastroenterology 2014;146:257-67. [Crossref] [PubMed]
- Baumgart S, Chen NM, Siveke JT, et al. Inflammation-induced NFATc1-STAT3 transcription complex promotes pancreatic cancer initiation by KrasG12D. Cancer Discov 2014;4:688-701. [Crossref] [PubMed]
- Chen NM, Singh G, Koenig A, et al. NFATc1 Links EGFR Signaling to Induction of Sox9 Transcription and Acinar-Ductal Transdifferentiation in the Pancreas. Gastroenterology 2015;148:1024-1034.e9. [Crossref] [PubMed]
- Andricovich J, Perkail S, Kai Y, et al. Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors. Cancer Cell 2018;33:512-526.e8. [Crossref] [PubMed]
- Bednar F, Schofield HK, Collins MA, et al. Bmi1 is required for the initiation of pancreatic cancer through an Ink4a-independent mechanism. Carcinogenesis 2015;36:730-8. [Crossref] [PubMed]
- Benitz S, Straub T, Mahajan UM, et al. Ring1b-dependent epigenetic remodelling is an essential prerequisite for pancreatic carcinogenesis. Gut 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Mallen-St Clair J, Soydaner-Azeloglu R, Lee KE, et al. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev 2012;26:439-44. [Crossref] [PubMed]
- Roy N, Malik S, Villanueva KE, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev 2015;29:658-71. [Crossref] [PubMed]
- Livshits G, Alonso-Curbelo D, Morris JP 4th, et al. Arid1a restrains Kras-dependent changes in acinar cell identity. Elife 2018; [Crossref] [PubMed]
- Genovese G, Carugo A, Tepper J, et al. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature 2017;542:362-6. [Crossref] [PubMed]
- Shakya R, Reid LJ, Reczek CR, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 2011;334:525-8. [Crossref] [PubMed]
- Shakya R, Gonda T, Quante M, et al. Hypomethylating therapy in an aggressive stroma-rich model of pancreatic carcinoma. Cancer Res 2013;73:885-96. [Crossref] [PubMed]
- Feldmann G, Karikari C, dal Molin M, et al. Inactivation of Brca2 cooperates with Trp53(R172H) to induce invasive pancreatic ductal adenocarcinomas in mice: a mouse model of familial pancreatic cancer. Cancer Biol Ther 2011;11:959-68. [Crossref] [PubMed]
- Skoulidis F, Cassidy LD, Pisupati V, et al. Germline Brca2 heterozygosity promotes Kras(G12D) -driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell 2010;18:499-509. [Crossref] [PubMed]
- Rowley M, Ohashi A, Mondal G, et al. Inactivation of Brca2 promotes Trp53-associated but inhibits KrasG12D-dependent pancreatic cancer development in mice. Gastroenterology 2011;140:1303-1313.e1-3.
- Li M, Chen Q, Ma T, et al. Targeting reactive nitrogen species suppresses hereditary pancreatic cancer. Proc Natl Acad Sci U S A 2017;114:7106-11. [Crossref] [PubMed]
- Mace TA, Shakya R, Pitarresi JR, et al. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer. Gut 2018;67:320-32. [Crossref] [PubMed]
- Russell R, Perkhofer L, Liebau S, et al. Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat Commun 2015;6:7677. [Crossref] [PubMed]
- Perkhofer L, Schmitt A, Romero Carrasco MC, et al. ATM Deficiency Generating Genomic Instability Sensitizes Pancreatic Ductal Adenocarcinoma Cells to Therapy-Induced DNA Damage. Cancer Res 2017;77:5576-90. [Crossref] [PubMed]
- Pinho AV, Van Bulck M, Chantrill L, et al. ROBO2 is a stroma suppressor gene in the pancreas and acts via TGF-beta signalling. Nat Commun 2018;9:5083. [Crossref] [PubMed]
- Blasco RB, Francoz S, Santamaria D, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 2011;19:652-63. [Crossref] [PubMed]
- Karreth FA, Frese KK, DeNicola GM, et al. C-Raf is required for the initiation of lung cancer by K-Ras(G12D). Cancer Discov 2011;1:128-36. [Crossref] [PubMed]
- Jiang SH, Li J, Dong FY, et al. Increased Serotonin Signaling Contributes to the Warburg Effect in Pancreatic Tumor Cells Under Metabolic Stress and Promotes Growth of Pancreatic Tumors in Mice. Gastroenterology 2017;153:277-291.e19. [Crossref] [PubMed]
- Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res 2005;65:3980-5. [Crossref] [PubMed]
- Sinha S, Fu YY, Grimont A, et al. PanIN Neuroendocrine Cells Promote Tumorigenesis via Neuronal Cross-talk. Cancer Res 2017;77:1868-79. [Crossref] [PubMed]
- Saloman JL, Albers KM, Li D, et al. Ablation of sensory neurons in a genetic model of pancreatic ductal adenocarcinoma slows initiation and progression of cancer. Proc Natl Acad Sci U S A 2016;113:3078-83. [Crossref] [PubMed]
- Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996;271:350-3. [Crossref] [PubMed]
- Yachida S, White CM, Naito Y, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res 2012;18:6339-47. [Crossref] [PubMed]
- Ijichi H, Chytil A, Gorska AE, et al. Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J Clin Invest 2011;121:4106-17. [Crossref] [PubMed]
- Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS Lett 2012;586:1959-70. [Crossref] [PubMed]
- Oshimori N, Oristian D, Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 2015;160:963-76. [Crossref] [PubMed]
- David CJ, Huang YH, Chen M, et al. TGF-beta Tumor Suppression through a Lethal EMT. Cell 2016;164:1015-30. [Crossref] [PubMed]
- Avila JL, Kissil JL. Notch signaling in pancreatic cancer: oncogene or tumor suppressor? Trends Mol Med 2013;19:320-7. [Crossref] [PubMed]
- Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer 2011;11:338-51. [Crossref] [PubMed]
- Miyamoto Y, Maitra A, Ghosh B, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 2003;3:565-76. [Crossref] [PubMed]
- Greer RL, Staley BK, Liou A, et al. Numb regulates acinar cell dedifferentiation and survival during pancreatic damage and acinar-to-ductal metaplasia. Gastroenterology 2013;145:1088-1097.e8. [Crossref] [PubMed]
- Avila JL, Troutman S, Durham A, et al. Notch1 is not required for acinar-to-ductal metaplasia in a model of Kras-induced pancreatic ductal adenocarcinoma. PLoS One 2012;7:e52133 [Crossref] [PubMed]
- Plentz R, Park JS, Rhim AD, et al. Inhibition of gamma-secretase activity inhibits tumor progression in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology 2009;136:1741-9.e6. [Crossref] [PubMed]
- Cook N, Frese KK, Bapiro TE, et al. Gamma secretase inhibition promotes hypoxic necrosis in mouse pancreatic ductal adenocarcinoma. J Exp Med 2012;209:437-44. [Crossref] [PubMed]
- Krop I, Demuth T, Guthrie T, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol 2012;30:2307-13. [Crossref] [PubMed]
- Tolcher AW, Messersmith WA, Mikulski SM, et al. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J Clin Oncol 2012;30:2348-53. [Crossref] [PubMed]
- Kopinke D, Brailsford M, Pan FC, et al. Ongoing Notch signaling maintains phenotypic fidelity in the adult exocrine pancreas. Dev Biol 2012;362:57-64. [Crossref] [PubMed]
- Hosokawa S, Furuyama K, Horiguchi M, et al. Impact of Sox9 dosage and Hes1-mediated Notch signaling in controlling the plasticity of adult pancreatic duct cells in mice. Sci Rep 2015;5:8518. [Crossref] [PubMed]
- Gao J, Azmi AS, Aboukameel A, et al. Nuclear retention of Fbw7 by specific inhibitors of nuclear export leads to Notch1 degradation in pancreatic cancer. Oncotarget 2014;5:3444-54. [Crossref] [PubMed]
- Pérez-Mancera PA, Rust AG, van der Weyden L, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 2012;486:266-70. [Crossref] [PubMed]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 2006;127:469-80. [Crossref] [PubMed]
- Serra S, Chetty R. Rnf43. J Clin Pathol 2018;71:1-6. [Crossref] [PubMed]
- Jiang X, Hao HX, Growney JD, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A 2013;110:12649-54. [Crossref] [PubMed]
- Steinhart Z, Pavlovic Z, Chandrashekhar M, et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat Med 2017;23:60-8. [Crossref] [PubMed]
- Sugimura R, Li L. Noncanonical Wnt signaling in vertebrate development, stem cells, and diseases. Birth Defects Res C Embryo Today 2010;90:243-56. [Crossref] [PubMed]
- Morris JP 4th, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer 2010;10:683-95. [Crossref] [PubMed]
- Chen NM, Neesse A, Dyck ML, et al. Context-Dependent Epigenetic Regulation of Nuclear Factor of Activated T Cells 1 in Pancreatic Plasticity. Gastroenterology 2017;152:1507-1520.e15. [Crossref] [PubMed]
- Singh SK, Chen NM, Hessmann E, et al. Antithetical NFATc1-Sox2 and p53-miR200 signaling networks govern pancreatic cancer cell plasticity. EMBO J 2015;34:517-30. [Crossref] [PubMed]
- Lee JS, Shukla A, Schneider J, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 2007;131:1084-96. [Crossref] [PubMed]
- McDaniel SL, Strahl BD. Shaping the cellular landscape with Set2/SETD2 methylation. Cell Mol Life Sci 2017;74:3317-34. [Crossref] [PubMed]
- Simon JM, Hacker KE, Singh D, et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 2014;24:241-50. [Crossref] [PubMed]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 2006;6:846-56. [Crossref] [PubMed]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011;11:481-92. [Crossref] [PubMed]
- Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013;45:592-601. [Crossref] [PubMed]
- Morel D, Almouzni G, Soria JC, et al. Targeting chromatin defects in selected solid tumors based on oncogene addiction, synthetic lethality and epigenetic antagonism. Ann Oncol 2017;28:254-69. [PubMed]
- Roberts NJ, Norris AL, Petersen GM, et al. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. Cancer Discov 2016;6:166-75. [Crossref] [PubMed]
- Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110-20. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. Erratum in: Nature 2013;502:258 Imielinsk, Marcin [corrected to Imielinski, Marcin]. [Crossref] [PubMed]
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 2011;12:68-78. [Crossref] [PubMed]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003;421:499-506. [Crossref] [PubMed]
- Cremona CA, Behrens A. ATM signalling and cancer. Oncogene 2014;33:3351-60. [Crossref] [PubMed]
- Huo Y, Cai H, Teplova I, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov 2013;3:894-907. [Crossref] [PubMed]
- Bowman-Colin C, Xia B, Bunting S, et al. Palb2 synergizes with Trp53 to suppress mammary tumor formation in a model of inherited breast cancer. Proc Natl Acad Sci U S A 2013;110:8632-7. [Crossref] [PubMed]
- Brose K, Bland KS, Wang KH, et al. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 1999;96:795-806. [Crossref] [PubMed]
- Long H, Sabatier C, Ma L, et al. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 2004;42:213-23. [Crossref] [PubMed]
- Mehlen P, Delloye-Bourgeois C, Chedotal A. Novel roles for Slits and netrins: axon guidance cues as anticancer targets? Nat Rev Cancer 2011;11:188-97. [Crossref] [PubMed]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009;136:701-18. [Crossref] [PubMed]
- Seiler M, Peng S, Agrawal AA, et al. Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell Rep 2018;23:282-296.e4. [Crossref] [PubMed]
- Krymskaya VP. Tumour suppressors hamartin and tuberin: intracellular signalling. Cell Signal 2003;15:729-39. [Crossref] [PubMed]
- Wang Y, Gogol-Doring A, Hu H, et al. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO Mol Med 2013;5:1431-42. [Crossref] [PubMed]
- Johnston JJ, Teer JK, Cherukuri PF, et al. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am J Hum Genet 2010;86:743-8. [Crossref] [PubMed]
- Chen Y, LeBleu VS, Carstens JL, et al. Dual reporter genetic mouse models of pancreatic cancer identify an epithelial-to-mesenchymal transition-independent metastasis program. EMBO Mol Med 2018; [Crossref] [PubMed]
- Cheung PF, Neff F, Neander C, et al. Notch-Induced Myeloid Reprogramming in Spontaneous Pancreatic Ductal Adenocarcinoma by Dual Genetic Targeting. Cancer Res 2018;78:4997-5010. [Crossref] [PubMed]
- Chiou SH, Winters IP, Wang J, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev 2015;29:1576-85. [Crossref] [PubMed]
- Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet 2018;19:671-87. [Crossref] [PubMed]
- Huch M, Bonfanti P, Boj SF, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 2013;32:2708-21. [Crossref] [PubMed]
- Boj SF, Hwang CI, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324-38. [Crossref] [PubMed]
- Tiriac H, Bucobo JC, Tzimas D, et al. Successful creation of pancreatic cancer organoids by means of EUS-guided fine-needle biopsy sampling for personalized cancer treatment. Gastrointest Endosc 2018;87:1474-80. [Crossref] [PubMed]
- Tiriac H, Belleau P, Engle DD, et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov 2018;8:1112-29. [Crossref] [PubMed]
- Gendoo DMA, Denroche RE, Zhang A, et al. Whole genomes define concordance of matched primary, xenograft, and organoid models of pancreas cancer. PLoS Comput Biol 2019;15:e1006596 [Crossref] [PubMed]
- Romero-Calvo I, Weber CR, Ray M, et al. Human Organoids Share Structural and Genetic Features with Primary Pancreatic Adenocarcinoma Tumors. Mol Cancer Res 2019;17:70-83. [Crossref] [PubMed]
- Huang L, Holtzinger A, Jagan I, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 2015;21:1364-71. [Crossref] [PubMed]
- Hou S, Tiriac H, Sridharan BP, et al. Advanced Development of Primary Pancreatic Organoid Tumor Models for High-Throughput Phenotypic Drug Screening. SLAS Discov 2018;23:574-84. [Crossref] [PubMed]
- Öhlund D, Handly-Santana A, Biffi G, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017;214:579-96. [PubMed]
- Biffi G, Oni TE, Spielman B, et al. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFbeta to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discov 2019;9:282-301. [Crossref] [PubMed]
- Chand S, O'Hayer K, Blanco FF, et al. The Landscape of Pancreatic Cancer Therapeutic Resistance Mechanisms. Int J Biol Sci 2016;12:273-82. [Crossref] [PubMed]
- Swayden M, Iovanna J, Soubeyran P. Pancreatic cancer chemo-resistance is driven by tumor phenotype rather than tumor genotype. Heliyon 2018;4:e01055 [Crossref] [PubMed]
Cite this article as: Tsuda M, Fukuda A, Takaori K, Seno H. Genetics and biology of pancreatic cancer and its precursor lesions: lessons learned from human pathology and mouse models. Ann Pancreat Cancer 2019;2:15.


