
Targeting the epigenome of pancreatic cancer for therapy: challenges and opportunities
Introduction
Pancreatic adenocarcinoma (PDAC) is currently the fourth leading cause of cancer death and is expected to climb to the second leading cause of cancer mortality by 2030, with the highest incidence-to-mortality ratios of any histology (1). At diagnosis, only 20% of patients are surgical candidates with any chance of cure; however, even with adjuvant therapy, the majority will recur and the median survival is under 2 years in this group, despite recent progress (2). Most patients have advanced disease at diagnosis, with a dismal overall prognosis that has remained virtually unchanged for many decades. Patients refractory to first-line therapy have 7% expected 5-year survival (3) and limited therapeutic options (4). Therefore, there is a great need for novel and more effective treatment strategies in PDAC.
The term epigenetics was first used by Waddington in 1942, was defined as “any heritable trait not involving the DNA sequence that influences the phenotype of a developing organism, providing a rapid and dynamic response to environmental changes”(5). Epigenetic refers to the somatically heritable differences in gene expression not attributable to intrinsic alterations in the primary sequence of DNA (6,7) (Figure 1) but to specific covalent modifications of chromatin components—which include DNA, RNA and proteins (e.g., histones). The vast majority of human cancers harbour both genetic and epigenetic abnormalities, with a fascinating interplay between the two. At present, the most studied epigenetic alterations associated with neoplastic phenotype include DNA methylation, histone modifications, and gene regulation by non-coding RNAs (8-11). In cancer, these epigenetic pathways can lead to silencing of important tumor suppressor genes or cell cycle checkpoints as well as hyperactivation of oncogenes and growth signaling pathways (12,13), contributing to cancer development and propagation (14-16).
Pathogenesis of PDAC has been most studied in the context of DNA mutations, suggesting potential therapeutic approaches targeting the molecular pathways disrupted by these mutations (17,18). However, genetic-based drivers of PDAC do not account for all of the phenotypic and molecular alterations. The identification of aberrant activated epigenetic pathways seen in early PanIN lesions through the development of PDAC strongly suggest that PDAC initiation and progression is the result of epigenetic changes that occurs in parallel to genetic ones, widening the window for therapeutic opportunity in PDAC (19-22).
In this review, we will discuss the epigenetic aberrations in PDAC and will review translational significance for the treatment of PDAC patients, discussing existing challenges and emerging strategies to overcome them.
DNA methylation
DNA methylation commonly occurs on extended regions of cytosine-guanine dinucleotides-called CpG islands- in the promoter regulatory regions of genes, resulting in the addition of a methyl group to the number 5 carbon of the cytosine pyrimidine ring to form 5-methylcytosine (6,23). Unmethylated CpG islands leads to an open chromatin state that allows gene transcription. The addition of the methyl group induces transcriptional silencing by interfering with transcription factors’ binding and recruiting methyl-CpG-binding domain proteins to initiate chromatin compaction (23,24). DNA methylation is catalyzed by a family of enzymes called DNA methyltransferases (DNMTs), responsible for maintenance and addition of methylation patterns, which can be either inherited or de novo modifications. DNMT1 functions mainly to maintain methylation patterns from the parent strand of DNA to the newly synthesized strand; DNMT3a and DNMT3b are responsible for de novo methylation (24,25). The ten eleven translocation (TET) proteins have been shown to mediate the conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian cells: this modification can be crucial as certain proteins, including DNMT1 do not recognize the 5-hydroxymethylcytosines, resulting in a loss of maintenance of methylation patterns (22).
Global demethylation may increase genomic instability and mutation rate (26); inappropriate methylation of the promoter region of genes can result in inactivation and silencing of genes critical for cell proliferation, DNA damage repair and apoptosis. Both process favor ultimately tumorigenesis (27).
DNMTs are over-expressed in about 80% of PDAC (28) and many promoters of tumor-suppressor genes are hypermethylated in PDAC (29-31). One example is the tumor-suppressor gene p16. The p16 protein inhibits the binding of the D-family cyclins to their cyclin-dependent kinase (CDK) partners, and its loss results in increased phosphorylation of retinoblastoma protein and subsequent progression through G1 phase into S phase of the cell cycle (32). Greater than 95% of PDACs have a loss of p16, with methylation of its promoter being one of the most common mechanisms of inactivation (32).
The use of genome-wide methylation analysis has allowed the identification of other genes affected by aberrant methylation in PDAC vs. normal pancreas (33), including genes involved in stem cell pluripotency (BM11, BMP3), WNT signalling (WNT5A, APC2, SOX1), cell adhesions (CDH2, CDH4) (34-36) and in the axonal guidance signalling pathway (SLIT/ROBO), which has been related with tumor neoangiogenesis (37). Additionally, the hypermethylation of the promoter regions of mismatch repair genes (hMLHI), growth inhibitory genes (ARHI), cell cycle control genes (cyclin D2), and proapoptotic genes (TNFRSF10c37, TMS138, and CRABP239) have also been reported in PDAC.
Yet it is important to note that in PDAC the loss of methylation of a normally silenced promoter is important as well. An example is VAV1, the gene encoding the hematopoietic-specific guanine nucleotide exchange factor: its promoter is demethylated in PDACs, leading to the activation of KRAS pathway expression and favoring cellular proliferation (38). Although there is an overall good degree of overlap in genome-wide studies about the set of genes found to be differently methylated in PDAC, some discrepancy exists (39-41). This variance could be explained by the different samples used (cell lines versus human tumor samples) as well as by analysis on varied platforms and how comprehensive the methylation analysis is. Nonetheless, these studies have been crucial to understand the significance of DNA methylation aberrations in driving PDAC, contributing to PDAC development and progression as well as to chemotherapy resistance.
Therapeutic significance of epigenetic alterations
DNMTs have been used as novel cancer therapeutic targets mostly due to their robust responses to inhibitors, credited to the intrinsic reversible nature of the methylation marks. The pyrimidine analogs, 5-azacytidine (AZA) and its deoxy derivative, 5-aza-2'-deoxycytidine (decitabine- DAC), are the most widely used DNMT inhibitors. When used at higher doses, these agents are also cytotoxic, owing to their direct incorporation into both DNA and RNA (AZA) or DNA only (DAC) (42,43). This strategy has already been shown to be effective in hematologic malignancies and AZA and DAC, have been FDA approved for the treatment of myelodysplastic syndromes and acute myeloid leukemia (44,45).
These DNA demethylating agents have also been used in preclinical model of PDAC. Kumari et al. showed that treatment of PDAC cell lines with AZA results in the reduction of telomerase activity, a key component of cellular immortality, and re-expression of the myeloid/lymphoid or mixed-lineage leukemia 3 (MLL3) gene, a tumor suppressor gene, usually downregulated in PDAC cell lines, tumor xenografts and archived patient tumors (46). A synergistic effect of AZA with either gemcitabine or docetaxel was also reported (47).
DAC treatment results in global DNA demethylation, tumor suppressor genes re-expression, significant growth inhibition and apoptosis of PDAC cell lines (48). Maitra and colleagues showed that while hypermethylation of the promoter regions of CRABP2 was associated with retinoic acid resistance in PDAC cell lines, treatment of these cells with DAC induced CRABP2 re-expression, increased apoptosis and restaured sensitivity to retinoic acid (49). Pan et al. showed a synergistic effect of emodin in combination with DAC in growth inhibition of Panc-1 cells, associated with enhanced demethylation of tumor-suppressor genes, such as p16, RASSF1A (50). Pretreatment with DAC increased the sensitivity of these cells to other chemotherapy agents, including gemcitabine and MEK inhibitors (51) (52). Recently, our group reported the chemosensitization effect of guadecitabine, a DAC prodrug with a favorable pharmacokinetic profile, in PDAC models. In an in vitro study, we showed that nanomolar doses of guadecitabine significantly improved the effects of irinotecan on decreasing cell viability, including in Panc1 model, which is usually non-responsive to either agent alone. From a mechanistic point of view, our results showed that guadecitabine functioned as a DNMTi with a `memory effect’ observed after 5-days of rest, followed by increased caspase 3/7 and LDH activity, thus suggesting that the optimal time for chemotherapy administration could be 5 days post-guadecitabine treatment (53).
The evidence of methylome dysregulation of PDAC has served as foundation for the development of clinical trials employing DNMTs inhibitors alone or in combination with chemotherapy or radiation as treatment for PDAC, and two phase 1 trials of AZA and nab-paclitaxel (NCT01845805) or gemcitabine (NCT01167816) are now ongoing in the metastatic setting. An orally active formulation of AZA, CC-486, has been developed providing a much more desirable route of administration as compared to the subcutaneous delivery of AZA. In the phase I study in patients with myelodysplastic syndromes, chronic myelogenous leukemia or refractory acute myeloid leukemia, CC-486 has proved to be bioavailable, well tolerated with similar response as the subcutaneous version. An investigator initiated, randomized phase II trial, of CC-486 in high risk, resected PDAC patients in now ongoing, with the main goal to decrease recurrence and/or improve response to chemotherapy at the time of recurrence (NCT01845805).
A study by Nagaraju et al. investigated the effect of curcumin analogues, EF31 and UBS109, as DNMT1 inhibitors in PDAC cells lines. Treatment resulted in re-expression of CDKN2A and p16 and a significant tumor growth inhibition, and increased the sensitivity to the combination of 5FU and oxaliplatin. However, the first-in-patient study assessing the feasibility of curcumin in combination with gemcitabine in advanced PDAC patients was associated with dose-limiting GI toxicity (54). In an attempt to reduce the toxicity, a nanoparticle-based curcumin (Theracurmin) has also been developed (55). In a phase I/II study evaluated the combination with gemcitabine in gemcitabine-resistant patients, although no increased toxicity from curcumin was reported, no responses were observed (55). Dhillon et al. conducted a subsequent monotherapy trial with curcumin in patients with advanced, pretreated PDAC. In this phase II trial, 25 patients received 8 g curcumin daily, which was well tolerated and showed biological activity with stable disease in one patient lasting over 18 months, and a brief but marked, tumor regression in another (56).
Important caveats related to DNMTs inhibitors should be acknowledged. First, DNA methylation is essential for a number of significant physiological pathways and global methylome dysregulation by these drugs may result in important toxicities, underscoring the impellent need to optimize their use. Second, hypomethylating drugs are S phase-dependent and both AZA and DAC have short half-lives, and therefore have low incorporation into DNA in many solid tumors (57-60). Finally, even in myeloid malignancies, primary and secondary resistance to these therapies are common and these drugs are most active when used as frontline therapy, a strategy that has never been investigated in PDAC (58,61).
Post-translational histone modifications
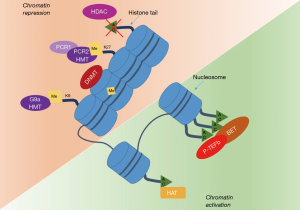
Histone modifications are cardinal component of the regulation of gene expression and constitute a dynamic process that is usually carried out by pairs of enzymes with reverse catalytic functions, essential for maintaining normal cellular function. The concept of “histone code” has indeed emerged to refer to histone-based regulation of gene transcription.
Most histone modifying enzymes act only on one or a select few histone marks to either place (writers), or remove (erasers) the mark on the histone tail or recognize the specific modification (readers) (12,62). Together with DNA methylation, these histone alterations have a pivotal role in regulating transcription, as each histone mark can recruit specific protein complexes that can either express or repress gene transcription. Acetylation of histone tails on lysine residues, mediated by histone acetyltransferases (HATs), changes the secondary structure of chromatin allowing transcription factors’ access to gene promoters. Deacetylation, performed by histone deacetylases (HDACs) enzymes, decreases transcription (11,63). Histone methylation, mediated by histone methyltransferases (HMTs), can occur on lysine, arginine and histidine residues, with lysine (K) methylation being the most characterized process: methylation of K9 and K27 in the histone tail of H3 induces the formation of heterochromatin, with subsequent transcriptional silencing (64,65). Additionally, each residue can be mono-, di-, or tri-methylated, providing another layer of regulation (65).
Current evidence suggests that aberrant histone modification patterns are critically involved in the PDAC tumorigenic process (66-69) and can help define subsets of patients with distinct epigenetic phenotypes and clinical outcomes. A study of three histone marks, H3 lysine 4 dimethylation (H3K4me2), H3 lysine 9 dimethylation (H3K9me2), and H3 lysine 18 acetylation (H3K18ac) suggested their role as independent predictors of poor survival in resectable PDAC patients (70). Enhancer of Zeste Homolog 2 (EZH2), a H3K27 methyltransferase and a component of polycomb protein complex (PRC2), is overexpressed in approximately 68% of PDAC (43). Nuclear localization and high expression of EZH2 is associated with poor-differentiation and shorter survival in metastatic setting.
Histone methylation
The most widely studied histone modifications are lysine alterations, including methylation, which is generally associated with transcriptional activation when occurring in promoter regions (71). G9a, is one of the main writers of this mark and has been found to be overexpressed in PDAC. G9a catalyzes the methylation of histone H3 at lysines 9 and 27 (H3K9 and K27) and works in complex with G9a-like protein (GLP) as well as with a variety of other epigenetic writers such as PRC2 (72,73).
Pharmacologic inhibition of G9a results in decreased H3K9 methyl marks, decreased proliferation via G2/M cell cycle arrest, and increased cellular senescence in pancreatic and other cancer cell models. G9a inhibition is even more effective as part of a combination therapy to revert chemoresistance, becoming a promising target for therapeutics (72). Pen and colleagues showed that inhibition of G9a using UNC0638 increased sensitivity to gemcitabine (74). These authors also showed that G9a expression correlated with the expression of the stemness genes including CD133, nestin and Lrg5 and its inhibition attenuates cancer stemness in these models. These findings were validated in vivo, where the combination treatment with G9a inhibitor and gemcitabine decreased tumor growth, lymph node invasion and distant metastasis.
Another histone tail modification frequently dysregulated in PDAC is hallmark H3K27 mono- and tri-methylation, mediated by EZH2, the catalytic subunit of PRC2. Drugs targeting EZH2 (UNC1999, GSK126) have shown promising results in PDAC models, (monolayer cells culture, spheroids, organoids and in vivo patient-derived xenograft mouse models), being associated with reduced aberrant K27 methylation, re-expression of cell cycle regulator p27Kip1, as well decreased PDAC cells proliferation rates, angiogenesis and increased apoptosis (75-77).
Ougolkov et al. noted an increase in the nuclear accumulation of EZH2 in chemo-resistant pancreatic tumor cells and showed that reversal of H3K27 methylation restored sensitivity to chemotherapeutics such as doxorubicin and gemcitabine (78). Co-exposure of DZNep (EZH2 inhibitor) and gemcitabine induced selective cytotoxic additivity in well- and poorly-differentiated PDAC cell lines, without affecting normal human pancreatic ductal epithelial cells (79). Interestingly, authors were able to show that a short priming with DZNep followed by gemcitabine treatment produced the maximal chemosensitization response (80).
Mathison et al. tested a combination of Aurora kinase A (AURKA) oncogene inhibitor and H3K9 methyltransferases inhibitor in vitro and in vivo models of PDAC, and showed that the combined inhibition of a genetic-to-epigenetic pathway was efficacious. Mechanistically, they reported that inhibition of H3K9 methyltransferases after targeting AURKA, arrested cells in G2-M phase, triggered an aberrant mitotic checkpoint response, and ultimately mitotic catastrophe (76). These data support the pathobiological hypothesis that PDAC develops and progresses in response to an interaction between known oncogenes and downstream epigenomic regulators. Increased interest has indeed emerged in the use of H3K27 methyltransferase inhibitors as part of combination therapies to re-sensitize resistant cells lines.
Histone deacetylation
Increased activity of HDAC is common in PDAC and lead to decreased histone acetylation and consequent gene repression. Jiao and colleagues, showed higher levels of HDAC3 protein expression in PDAC tissues and cell lines as compared to paired normal ones, associated with cell proliferation, migration and invasion, and increase drug resistance (81).
HDAC2 and HDAC7 expression also increased in PDACs, especially in poorly-differentiated cases. Increased expression of HDAC7 can distinguish PDAC from other benign pancreatic neoplasms and is associated with apoptotic resistance of cancer cells via silencing of the apoptotic-inducing NOXA gene and attenuation of TRAIL-induced apoptosis (82-84). HDAC1 is found at increased levels in the vast majority of PDACs and its precursor lesions. HDAC1 and HDAC2/SIN3a are recruited to the TGFBR2 promoter leading to repressed expression of this tumor suppressor gene, in the absence of any genetic alterations. HDAC1/2-mediated transcriptional control regulates the epithelial-mesenchymal transition in PDAC cells, thereby contributing to invasion and metastasis (85).
As a result of the frequently observed dysregulation of HDAC family members in PDAC, and their role in controlling key oncogenic features, inhibition of HDAC has been investigated as potential therapy for PDAC patients. Several natural and synthetic compounds that inhibit HDAC activity are now available, which either target all HDAC family members (pan-HDAC inhibitor) or selectively interfere with subgroups of HDAC isoforms, with interesting activity shown in preclinical models (13,86).
Knockdown of HDAC3 gene through lentivirus-mediated methods inhibits PDAC cells proliferation and enhances the sensitivity to gemcitabine treatment, consistent with the effect of HDAC inhibitor (HDACi) trichostatin A and 4-phenylbutyrate (TSA) (87-90). In PDAC cell lines, treatment with HDACi vorinostat induces growth inhibition and G1 cell cycle arrest via upregulation of p21, with an additive effect on growth inhibition when combined with gemcitabine. Fritsche et al. noted that HDAC2 is upregulated in PDAC cells that acquire resistance to etoposide and treatment with HDACi, valproic acid, in combination with etoposide increased apoptosis and restored the etoposide sensitive phenotype in resistant cells (83).
In the clinical setting, monotherapy treatment with HDACi has showed activity in hematological malignancies, with vorinostat, romidepsin and panobinostat reaching approval for the treatment of cutaneous T-cell lymphoma (vorinostat and romidepsin) (91), peripheral T-cell lymphoma (romidepsin) (92) and multiple myeloma (vorinostat and panobinostat) (93). However, although effective in cell lines, HDAC inhibitors in monotherapy have not shown efficacy in early phase clinical trials for PDAC.
Consequently, clinical investigations have been focused on combined approaches with small-molecule inhibitors, chemotherapeutic or immunotherapy agents, with the intent to use HDAC inhibition to manipulate the microenvironment of the tumor to increase the sensitivity to standard therapeutics. A phase I trial tested oral panobinostat combined with gemcitabine in advanced solid tumor patients, including three PDAC patients, of which one had stable disease under treatment (94). A similar trial investigating the combination of mocetinostat with gemcitabine in patients with advanced PDAC was early terminated because of lack of efficacy combined with significant toxicity profile (95).
Romidepsin has been evaluated in combination with gemcitabine in solid tumors, including PDAC. No responses were observed in the ~25% of individuals with PDAC, associated with additive hematological toxicity of the combination (96).
The pan-HDAC inhibitor belinostat has been shown to decrease growth and increase apoptosis in PDAC preclinical models, mainly via blocking the AKT/mTOR pathway. In a phase I clinical trial evaluating belinostat in combination with carboplatin and/or paclitaxel in patients with solid tumors, one partial response was observed among the three PDAC patients enrolled, but as platinums and taxanes have activity in PDAC, it is uncertain what the contribution of the HDACi was (97). In two phase I studies testing the safety and activity of entinostat given alone and in combination with 13-cis-retinoic acid (13-cR), one PDAC patient either was enrolled, achieving stable disease in the first setting, while progressive disease was noted after combined treatment (98). Similarly, a phase I/II study has evaluated the toxicity and efficacy of valproic acid in combination with S-1 in pancreatobiliary patients, reported clinically significant agent-related adverse events in 67% of the patients enrolled, including G3–4 anemia and thrombocytopenia (99).
No objective responses were reported when vorinostat was added to chemoradiation with capecitabine in a phase I dose-finding study of 21 patients with non-metastatic PDAC, although 90% had stable disease (100).
These studies revealed minimal effects in a limited number of PDAC patients for the combination of HDACi as compared to gemcitabine monotherapy (101). Moreover, in all these studies high grade, treatment-related myelosuppression and gastrointestinal events were common, raising concern about the safety profile of this approach.
As an alternative approach, several trials have been evaluated the combination of HDACi and targeted therapies in PDAC. Studies in vitro in PDAC showed a synergistic effect of the combination of the proteasome inhibitor marizomib and vorinostat; however, no responses were found in a phase I clinical trial using this strategy (102). Similarly, no efficacy was shown for the combination of panobinostat with the proteasome inhibitor, bortezomib, in a phase II clinical trial in gemcitabine-resistant PDAC patients, with high rate of grade 3 and 4 toxicities reported (thrombocytopenia and diarrhea) (103).
An ongoing neoadjuvant clinical trial is now investigating the efficacy of vorinostat and sorafenib plus standard therapy (gemcitabine plus nab-paclitaxel or gemcitabine plus radiation) and sorafenib in patients with stage I–III PDAC (NCT02349867).
Unfortunately, combinatory treatment regimens with strong and promising mechanistic synergism in preclinical PDAC models failed in first-in-patient studies, thus reflecting the difficulties of translating these findings into the clinical setting. One of the main difficulties of HDAC inhibition therapy is their global repressive effects with lack of target specificity: the outcome of these drugs is not predictable and can be associated with increased toxicity. Further careful investigations are highly needed to understand the impact of HDAC inhibition on both antitumor activity and toxicity
Nguyen et al. advanced the hypothesis that stromal fibroblasts can contribute to the poor efficacy of HDAC inhibition in PDAC. A key mechanism by which CAFs modify the behavior of neighboring tumor cells is via release of proinflammatory factors into the tumor microenvironment (TME) (104). These authors showed that HDACi-treatment of CAFs caused a dose dependent increase in the expression of inflammatory genes, causing a counter-productive and paradoxical tumor supportive phenotype. Combination therapies targeting PDAC stroma may mitigate these unintended effects and enhance their efficacy as anti-tumor drugs.
Finally, epigenetic mechanisms are intertwined as part of a broader spectrum of cellular mechanisms including DNA repair and DNA-damage signaling. Dynamic regulation of acetylation events on H3K56 and H4K16 and recruitment of HDAC1/2 to sites of DNA double-strand breaks (DSBs) have been reported (105).
Chen and colleagues showed that treatment of PDAC cells with the HDACi AR-42 induced ROS and caused DNA damage, particularly double-strand breaks (DSBs), leading to activation of both caspase-dependent and -independent apoptosis pathways. This was associated with decreased cell invasiveness in vitro and suppressed tumor growth in vivo (106). Agarwal et al. showed that the G9a inhibitor, UNC0638, sensitizes PDAC cells to DNA-damaging agents, by impairing DSBs repair (72). Both epigenetic and DNA repair pathways are aberrantly regulated in PDAC, especially in the subtypes carrying germline or sporadic mutations in BRCA genes (107). Targeting epigenetic and DNA repair pathways simultaneously might strongly impede cancer cell proliferation and provide new opportunities for future PDAC combination therapies.
Together, these results suggest that an optimized exposure to epigenetic modulatory drugs (EMD) can sensitize PDAC to other therapeutic agents, and emphasize the promising clinical utilities of epigenetic reversal agents in future PDAC combination therapies (Table 1).
Table 1
| Drug group | Drug name | Combination agent(s) | Clinical trial phase, disease stage | Results and best response in PDAC | Clinical trial Ref. |
|---|---|---|---|---|---|
| HDAC inhibitors | |||||
| HDAC Pan-inhibitors | Panobinostat | Bortezomib | 2, metastatic, gemcitabine-resistant PDAC | 3 patients with PDAC, all had PD and severe treatment-related toxicity, study was early closed | Wang et al. 2012 (103) |
| HDAC class I and II | Vorinostat | Marizomib | 1, metastatic PDAC | 3 patients with PDAC, all had PD | Millawrd et al., 2012 (102) |
| 5-FU + radiation | 1, non-metastatic PDAC | 19 patients had SD, and 2 had PD | Chan et al., 2016 (100) | ||
| Capecitabine + radiation | 1/2, locally advanced PDAC | Not available, study early terminated | NCT00948688 | ||
| Gemcitabine/Sorafenib +/− radiation | 1, non-metastatic PDAC | Ongoing and recruiting | NCT02349867 | ||
| Valproic Acid | Epirubicin | 1, advanced solid tumors | PR in one PDAC patient | Munster et al., 2007 (108) | |
| Gemcitabine + radiation | 2, locally advanced PDAC | Not available, study terminated | NCT01333631 | ||
| Romidepsin | Gemcitabine | 1, advanced solid tumors | SD in 6/10 patients with PDAC, treatment associated with significant TEAEs | Jones et al., 2012 (96) | |
| HDAC class I | Entinostat | 13-cis retinoic acid | 1, advanced solid tumor | One patient with PDAC enrolled had SD | Pili et al., 2012 (98) |
| FOLFOX | 1b, metastatic PDAC | Not yet recruiting | NCT03760614 | ||
| CI-994 | Gemcitabine | 2, randomized to gemcitabine advanced PDAC | Inferior in OR and survival compared to gemcitabine monotherapy with decreased quality of life | Richards et al., 2006 (101) | |
| HDAC class I + IV | Mocetinostat | Gemcitabine | 1/2, advanced solid tumors | Phase 1: 12 patients with PDAC, 2 had PR; |
Chan et al., 2018 (95) |
| HMT inhibitors | Curcumin | – | 2, metastatic PDAC | 21 evaluable PDAC, one patient had SD and one had PR | Dhillon et al., 2008 (56) |
| Gemcitabine | 1/2, advanced PDAC | 11 evaluable PDAC, 1 PR and 1 SD, but overall treatment discontinued very early due to toxicity | Epelbaum et al., 2010 (54) | ||
| Gemcitabine | 1/2, metastatic, gemcitabine resistant PDAC | 21 enrolled PDAC, 5 had SD | Kanai et al., 2011 (55) | ||
| DNMT inhibitors | CC-486 (oral azacitidine) | – | 2, high risk resected PDAC | Ongoing and recruiting | NCT01845805 |
| Gemcitabine | 1, advanced | Not available, study early terminated | NCT01167816 | ||
| Rx-3117 | nab-paclitaxel | 1,2 first line in PDAC | Ongoing and recruiting | NCT03189914 | |
| BET inhibitors | MK-8628 | – | 1, advanced solid tumors including PDAC | Completed, no results yet | NCT02259114 |
| Bay1238097 | – | 1, advanced solid tumors including PDAC | Study was prematurely terminated because of the occurrence of DLTs | Postel-Vinay et al., 2019 (109) | |
| BI-2536 | – | 2, advanced PDAC | 86 evaluable patients, 2 PR, but both patients discontinued treatment to clinical or non-target lesion progression, OS was 4.9 months | Mross et al., 2012 (110) | |
| INCB057643 | Monotherapy (part 1 and part 2) and in combination with standard-of-care (SOC) agents (part 3 and 4) | 1/2, advanced solid tumors including PDAC | Active, not recruiting | NCT02711137 |
BET, bromodomain and extra-terminal; DNMT, DNA methyltransferase; HDAC, histone deacetylase; HMT, Histone methyltransferases; PD, progressive disease; PDAC, pancreatic adenocarcinoma; PR, partial response; SD, stable disease; TEAEs, treatment emergent adverse events; EMD, epigenetic modulatory drug.
Histone acetylation
Acetylation of histones and non-histone proteins of specific lysine residues by HATs neutralizes the positive charge on the amino group, weakening the DNA-chromatin complex and creating an open chromatin configuration, which facilitates gene expression. One study evaluated H4K12 and H3K18 acetylation in PDAC patients’ samples by immunohistochemistry and found that these marks were indicators of lower overall survival (111). Decreased expression of p300, a HAT, has been reported in highly metastatic PDCA cell lines, supporting its role as a classical tumor suppressor protein (112). However, the role of histone acetylation in PDAC tumorigenesis and progression is still unclear, and highly dependent on the cellular context.
Careful preclinical investigations are still required and no current studies are investigating drugs targeting these proteins in this context.
Bromodomain epigenome readers and their inhibitors: a novel therapeutic target
While acetylation levels are regulated by HATs (“writers”) and HDACs (“erasers”), acetylation marks are recognized by bromodomain-containing proteins (“readers”), such as the bromodomain and extra-terminal motif (BET) family of chromatin adaptors (BRD2, BRD3, BRD4 and BRDT) (113). These proteins interact with the acetylated lysine residues of the histone tails to facilitate the recruitment of macromolecular transcription complexes necessary for the transcription of specific subset of genes, further enhancing the transcriptional activation resulting from the acetylation marks. These proteins also function as mediators of transcriptional elongation by promoting the recruitment and activation of the positive transcription elongation factor-b complex (P-TEFb) (114,115).
Human BRD4 was initially identified in NUT midline carcinoma (NMC), a rare subtype of squamous cell carcinoma characterized by a translocation most often involving the NUT gene and BRD4 (116). NMC, typically arising from the midline structures of the head, neck, and thorax, is extremely aggressive tumor, with median overall survival of 6.7 months (117). Treatment of NMC cells with BET inhibitors results in proliferation arrest in in vitro and in vivo models, and treatment with the oral BET inhibitor OTX015/MK-8628 led to significant and rapid tumor regression in 2 NMC patients (118). Now multiple BET inhibitors have shown clinical activity in NUT midline patients (119,120).
While initial reports suggested the transcriptional inhibition of oncogenic c-Myc as the crucial mechanism of BET inhibition antitumor activity (121,122), recent studies have shown the role of the BET proteins in various differentiation pathways and in controlling other cancer-relevant genes such as BCL2, FOSL154, as well as the activity of the EMT-related transcription factor Twist1. These findings underscore the potential of small-molecule inhibitors that specifically target these readers of for solid tumors’ treatment and tumor reprogramming (114,123,124). One of the founding BET inhibitor small molecules is JQ1, which was initially described to be a potent suppressor of NMC and B-cell lineage malignancies (115). Expression of BRD2, BRD3, and BRD4 has been detected in both preneoplastic lesions and PDAC and administration of JQ1 blocked acinar-to-ductal metaplasia—a key event in PDAC initiation—the development of PanINs lesions and PDAC cells proliferation. These effects were associated with decrease activation of the pro-survival kinase AKT and with downregulation of inflammatory regulators such as STAT3 and IL6 (125). Wang and colleagues showed that BRD4 was significantly upregulated in PDAC cell lines upon treatment with gemcitabine and combination treatment with BET inhibitors had a synergistic effect (126).
These data suggested that BET proteins play an important role in PDAC growth, progression and chemoresistance, making it a promising target for anticancer treatments in this disease (127). Sahai et al. demonstrated that JQ1 and BRD4 knockdown suppress proliferation of chemotherapy resistant- PDAC cells in an in vitro three-dimensional collagen model (128). Later, Garcia et al. using five PDAC patient-derived xenograft models, showed that JQ1 treatment was associated with significant tumor growth suppression. No consistent association with decreased c-Myc expression was observed, while significant inhibition of CDC25B expression, a regulator of cell cycle progression, was reported (129). A more recent study confirmed the antitumoral activity of BET inhibitors in PDAC human-derived xenograft tumors (127).
In terms of combination approaches, JQ1 in combination with gemcitabine led to a significant reduction in tumor volume and proliferation in a Kras;p53 mutant PDAC mouse model (125). Functional studies confirmed that BET inhibition alters PDAC’ TME by decreasing the protective stroma formed by CAF: inflammatory signals and expression of the tumor-associated stroma markers were all reduced upon JQ1 treatment, consistent with other reports (128,130,131), providing a unique example of simultaneous targeting of both the stromal and neoplastic cells.
Clinical trials using different BETi have been initiated in PDAC (NCT01987362, NCT02259114, and NCT02369029) (Table 1). However, first clinical studies testing BET inhibitors in monotherapy in PDAC have been discouraging: a randomized phase II trial in patients with unresectable PDAC using BI-2536, an inhibitor of the Polo-like kinase that has been shown to block BRD4 activity in vitro (NCT00710710), yielded poor response rates.
These data are not completely surprising: considering that JQ1 treatment had only a modest effect on survival on its own in preclinical models, paving the pathway for the use of these drugs not as a standalone treatment, but combination with other therapies. Mazur et al. showed that JQ1 synergizes with the HDAC inhibitor, vorinostat, in a KrasG12D;p53ko PDAC model. Similar antitumourigenic activity of BETi/HDACi treatment was shown in a preclinical model of lymphoma and acute myelogenous leukaemia and in KRAS mutant lung cancer models (125,132,133). Several mechanisms have been proposed to explain the antitumor activity of combined BETi/HDACi, including de-repression of p57, P-TEFb recruitment and subsequent transcriptional induction and elongation of a defined set of target genes.
In summary, BET proteins clearly play a role in PDAC pathology, but additional studies are required to optimize their value as therapeutic target and biological marker in PDAC.
Epigenetic regulation by noncoding RNAs
Non-coding RNAs (ncRNA) are RNAs that are not translated into proteins and include different classes of small RNAs [<200 bases, like microRNA (miRNA)] and long non-coding RNAs (lncRNAs, >200 bases), whose expression is tissue and stage specific. NcRNA are mainly implicated in translational repression and RNA degradation, but recent findings underscored their interaction with chromatin modifier complexes in gene regulation.
MiRNAs bind complementary regions of mRNAs, usually in the 3’ region, and inhibit the process of translation or decrease the stability of the associated mRNA specie. Each miRNA may have many mRNA targets. Numerous miRNAs are abnormally expressed in PDAC and its precursor lesions (134) and miRNA profiling has been shown to be effective in differentiating normal tissue from PDAC and could facilitate early diagnosis. Examples include miR-155 and miR-21. MiR-155 has been used in diagnosing IPMNs in pancreatic juice samples and its levels increase progressively in PanIN2 and PanIN3 lesions (135,136). Similarly, miR-21 expression increases with PanIN grade, with peak expression occurring in hyperplastic PanIN-1/2 lesions and when tested in pancreatic cyst fluid, was found to be an encouraging biomarker to differentiate cancer patients from those with chronic pancreatitis and healthy subjects (137-139).
Extension of these works demonstrated that a number of miRNAs are deregulated in patients with PDCA as compared to healthy controls (140,141). Meta-analyses of these studies have identified a few miRNAs that are reported in multiple studies as consistently altered in PDAC (i.e., miR-21 and miR-23a were identified as upregulated, and miR-148a and miR-375 as downregulated in multiple profiling studies), pointing out the complexity of the miRNA transcriptome (142,143).
MiRNAs levels are also associated with PDAC clinical outcomes. Increased expression of miR-21, miR-155 (140), miR-196a-2 (144), miR-203(145), and miR-183 (146) are associated with poor prognosis, while miR30a-3p, miR-105, miR-127, miR-187, miR-452, and miR-518a-2 predict better survival in PDAC patients with lymph node positive disease (140).
Other studies suggested that miRNAs modulate chemo-resistance to gemcitabine in PDAC (147-149). Treatment of PDAC gemcitabine-resistant cell lines with lentiviral vectors containing miR-181b mimics resulted in increased sensitivity to the drug. Similar findings were obtained in PDAC xenografts models (150). Wang et al., reported that miR-23b is downregulated in radioresistant PDAC cells and its restoration increases the sensitivity to radiation therapy (151).
LncRNA have also emerged as a major mechanism for PDAC tumorigenesis by regulating important cellular behaviors such as cell proliferation, invasion, metastasis and chemoresistance. LncRNAs are also potential diagnostic and prognostic biomarkers in PDAC (35,152). A recent genome-wide study showed that germline variation of lncRNA, LINC00673, might confer susceptibility in development of PDAC (153). LncRNA HOTAIR, PVT, H19 are overexpressed in PDAC and H19 has been associated with tumor grade and metastasis (154-156). Uc.345 is also upregulated in PDAC tissues and correlates with higher stage and decreased overall survival (157). Plasma levels of Linc-pint, a p53-induced lncRNA, are associated with higher risk tumor recurrence, and correlate with poor prognosis (158).
Therefore, ncRNAs are potential diagnostic, prognostic biomarkers of PDAC and there is significant enthusiasm about their role as therapeutic target (159). Restitution of miRNAs through nanoparticle delivery has been investigated preclinically in PDAC. The miR-34a nanocomplexes alone or combined with miR-143/145 nanovectors significantly suppressed the growth of gemcitabine resistant MiaPaCa-2 subcutaneous xenografts and orthotopic PDAC models (160,161). It has also been shown that the combination of gemcitabine with miR-205 is able to overcome drug resistance and inhibit invasion of gemcitabine resistant PDAC cells and animal models (162).
Another promising approach to target small ncRNAs involves antisense oligonucleotides, which function binding miRNAs with high complementarity to inhibit their function. Administration of the combination of anti-miR-21 and anti-miR-221 oligonucleotides significantly reduced tumor growth and metastasis in PDAC models (163). Similar findings were reported with inhibition of miR-132 and miR-212 by antisense miRNA (164).
Several natural agents including isoflavone, curcumin, 3,3'-diindolylmethane (DIM), have been investigated for their effects on the regulation of miRNAs in PDAC. Studies showed that in PDAC cells isoflavone could normalize the levels of several miRNA (including miR-27a, miR-146a, miR-200, miR-34a), resulting in suppressed cancer cell proliferation and invasion through the inactivation of Akt and NF-κB pathway. However, in a phase 2-study in patients with advanced PDAC, the addition of soy isoflavones to gemcitabine and erlotinib did not improve patients’ outcome, although the triplet appeared to be well tolerated (165). DIM and curcumin have also shown potential anti-cancer activities in PDAC through miRNA regulations. Treatment of MiaPaCa-2 and Panc-1 cells with either DIM or curcumin resulted in cell growth and migration inhibition via the down-regulation of miR-221 and subsequent induction of PTEN, p27, p57, and PUMA (166). Another phase 1/2a clinical trial investigated the role of intratumoral administration of BC-819 in locally advanced PDAC. BC-819 is a DNA plasmid carrying the gene for the diphtheria toxin- under the regulation of the lncRNA H19 gene promoter, which is overexpressed in PDAC. The maximum tolerated dose of BC-819 was not reached in this study and encouraging results were observed in term of tumor response (167).
Although, ncRNAs are promising therapeutic agents, therapeutic application is still in its infancy and to date there are no miRNA-based therapy approved for PDAC. Several challenges exist for their application, including in vivo instability and lack of gene targeting specificity. Additional pre-clinical and proof-of-concept clinical studies are required to better understand the meaning of these ncRNAs in PDAC and their possible value as therapeutic targets.
The impact of epigenetic therapeutics in pancreatic cancer immunity and immunotherapy
The understanding of the role that immune checkpoint molecules, such as cytotoxic T-cell lymphocyte-4 antigen (CTLA-4) receptor and programmed death-1 (PD-1) T cells co-receptor and its ligands PD-L1/PD-L2, play in the maintenance of immunosuppression within the TME has led to the clinical development of monoclonal antibodies targeting these molecules [immune checkpoint inhibitors (ICIs)] (168-170). ICIs have become a major focus of cancer therapy.
Antitumor immune response and therapy effectiveness depends on the ability of cytotoxic T cells (Teff) to infiltrate the tumor and recognize tumor cells and on the amount of tumor antigens and intact antigen presentation machinery (171). Cancer types with higher mutation burden, and consequently higher probability of neo-antigens, frequently show higher response rates to ICIs: in responsive patients, these immune checkpoint blockade therapies have resulted in long-term control of chemotherapy-resistant tumors that can last years (172,173).
The benefits are much more limited in non-immunogenic tumors lacking T cell infiltrate, such as PDAC, characterized by a hostile and immunosuppressive microenvironment that impedes T cell infiltration and function (174,175).
Recently, it has become apparent that EMD are capable of enhancing tumor immunogenicity and boosting the antitumor immune response, and several studies have already demonstrated synergy between immunotherapies and EMDs for cancer treatment (176). Multiple groups have demonstrated that the administration of low doses of DNMTIs, though not cytotoxic to cells, causes wide promoter DNA demethylation and reprogramming of regulatory pathways in tumor cells, increase expression of genes in the type I IFN, antigen processing and presentation, PD-1/PD-L1 pathways and induce the expression of cancer testis antigens in various cancer types (177,178). Other group showed that DNA hypomethylating agents and HDAC inhibitors can also reactivate endogenous retroviral elements (ERV), thus enhancing an intrinsic immune system response. Specifically, the long terminal repeats (LTRs) of ERVs, normally silenced by DNA methylation, can become re-expressed with these agents, leading to transcriptional expression of thousands of previously non-annotated transcription start sites and subsequent activation of an antiviral innate immune response and creating a state of ‘viral mimicry’ (179).
Recent work with HDACi has shown the ability of these agents to alter the immunogenicity of the TME by inducing the expression of tumor associated antigens, increasing tumor cell expression of MHC class II, inducing the expression of natural killer cell receptors and ligands and decreasing Tregs and myeloid-derived suppressor cells (MDSCs) in multiple different tumor models (180,181).
BETi also have intrinsic immunomodulatory properties that favor antitumor immunity. Zhu et al. were among the first to describe that CD274, the gene encoding PD-L1, was a direct target of BRD4; BETi directly suppressed PD-L1 transcription in cancer and immune cells (182). The potential for BET inhibitors to induce immunogenic cell death has also been suggested (183).
Peng et al., using a mouse model of ovarian cancer, reported increased expression of the Th1-type chemokines CXCL9 and CXCL10 upon EZH2 inhibition and DNMT1i exposure, resulting in increased T-cell trafficking in the TME, which enhanced therapeutic efficacy of PD-L1 checkpoint blockade as well as adoptive T-cell therapy (184). In preclinical models of renal and castration resistant prostate cancer, low dose of HDACi entinostat in combination with IL-2 therapy or a survivin based vaccine inhibited tumor growth, reduced infiltrating regulatory T cells (Tregs), and increased the Teff response (185). A synergistic effect of ICIs in combination with entinostat and AZA was reported also in preclinical models of CRC and breast cancer (186).
In NSCLC patients who previously underwent epigenetic therapy and subsequently began immune checkpoint therapy, all five patients passed the 24-week point without progression with three of these individuals maintaining partial RECIST responses for over 2.5 years (187). Other clinical trials combining the HDACi entinostat and the anti-PD-1 pembrolizumab in NSCLC and melanoma have demonstrated promising activity in anti-PD-L1-resistant patient groups. Of note, responses were observed even in the absence of PD-L1 expression by IHC (188). Multiple other phase I/II trials are underway testing DNMTi and/or HDACi in combination with anti-PD1 therapy in multiple histologies with results expected soon.
Acknowledgments about the precise relationships between epigenetic aberrations, immune system and the consequences for cancer cell phenotypes could have tremendous translational implications in PDAC. The TME in PDAC is remarkable for its profound desmoplasia and absence of Teffs and its T helper 2 cell immunophenotype (189). This allows PDAC to avoid immune surveillance and explains the ineffectiveness of ICIs in numerous studies of metastatic PDAC patients (190-192). Therefore, epigenetic modulation might represent a novel strategy to prime the tumor and TME and reverse immunosuppression in PDAC.
Shakya et al. tested DAC in an aggressive stroma-rich mouse model of PDAC and showed that DAC was able to slow disease progression and induce transient tumor growth inhibition. Furthermore, an additive antiproliferative effect on PDAC cells was reported for the combination of DAC plus IFN-γ (193), providing a rationale for future studies combining hypomethylating agents with cytokines and immunotherapy.
Lu and colleagues showed that CD274 promoter is enriched for H3K4 trimethylation (H3K4me3), catalyzed by MLL1 and result in PD-L1 transcription in PDAC cells. In an orthotopic mice model, they showed that inhibition of MLL1 decreases the H3K4me3 levels in the CD274 promoter and PD-L1 expression, and resulted in significant tumor growth suppression when combined with anti-PD-L1 or anti-PD-1 antibody (194).
Our laboratory has explored the role of HDAC inhibition in immunocompetent murine PDAC models, and we demonstrated that the HDACi, entinostat, shifted MDSCs from a myeloid-MDSC-dominant population to the less immunosuppressive G-MDSCs subtype. The functional capability of these cells was also impaired, with the remaining MDSCs expressing less Arginase-1 and less PD-L1 (195). Combination therapy of entinostat with anti-PD1 agent or anti-CTLA4 antagonist antibody significantly improved survival as compared to either agent alone. Based on these preclinical data, a phase 2 study evaluating the clinical efficacy of entinostat plus anti-PD-1 in unresectable or metastatic PDAC has been initiated (NCT03250273).
Several challenges need to be considered when developing epigenetic-immune therapy combinations, including the optimal drug sequence, whether the treatments should be sequential or concurrent, continuous or intermittent, and the optimal dose (196). However, multiple lines of evidence presented here suggest that exploiting cancer epigenome may result in increased immunogenicity and overcome resistance to immune therapies, supporting the hypothesis that EMD may prime an ICI insensitive cancer into a sensitive one, and clinical trial have been initiated (Table 2).
Table 2
| EMD class | EMD name | Immunotherapy target | IO agent name | Clinical trial phase, disease stage | Status | Clinical trial Ref. |
|---|---|---|---|---|---|---|
| HDAC inhibitor | Entinostat | PD-1 | Nivolumab | 2, advanced PDAC and cholangiocarcinoma | Ongoing, recruiting | NCT03250273 |
| DNMT inhibitor | Guadecitabine | PD-L1 | Durvalumab | 1b, advanced HCC, PDAC, cholangiocarcinoma | Ongoing, recruiting | NCT03257761 |
| DNMT inhibitor | Azacitidine | PD-1 | Pembrolizumab | 2, advanced PDAC | Ongoing, recruiting | NCT03264404 |
DNMT, DNA methyltransferase; EMD, epigenetic modulatory drug; HCC, hepatocellular carcinoma; HDAC, histone deacetylase; IO, immune-oncology; PDAC, pancreatic adenocarcinoma; PD-1, programmed cell death; PD-L1, programmed death-ligand.
Conclusions
The role of epigenetics in PDAC carcinogenesis is now better defined and has been linked to increased cancer cell stemness, altered cellular metabolism, differentiation, and chemoresistance (197). Unlike genetic alterations, epigenetic plasticity allows rapid, dynamic and potentially reversible changes that favor tumor growth and progression, as well as immune escape and resistance to therapies (7,13).
However, how exactly alterations in epigenome affect PDAC development and progression, is still not fully understood (41).
The overall satisfactory tolerance of epigenetic drugs with minimal overlapping toxicities with other classes of drugs, together with their intrinsic immunomodulatory effects, make them promising therapeutic approach to combine with conventional therapies and immune therapies.
However, there is no doubt that much still need to be done to optimize the use of epigenetic drugs before translating these agents into the clinical practice. The discrepancy noted between the promising preclinical data and the modest clinical efficacy reported in early phase clinical trials in PDAC patients raises important concerns (198). In the era of personalized medicine, a better understanding of which subset of patients could benefit most from certain EMD treatment is highly needed. Appropriate preclinical models should be used to explore the molecular rational of combinatorial regimens and set the stage for future clinical trials. The use of dedicated pharmacodynamics companion biomarkers in these studies may guide the determination of the optimal dosage, schedule and population. Accordingly, comprehensive translational studies should be carried out to determine whether the observed effect of EMDs in PDAC is linked to the effect of the drug on the tumor, on the stroma and/or on specific subsets of immune cells.
In conclusion, our increasing knowledge of the genetic and epigenetic aberration that drive PDAC pave the pathway for novel, promising and exciting therapies in this setting. Laboratory-based studies, clinical and translational studies are warranted to better understand the complex interactions of PDAC genetics, epigenetics and immunology to allow the translation of these findings into clinical practice.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Colin Weekes) for the series “Systemic and Targeted Therapies for Pancreas Ductal Adenocarcinoma” published in Annals of Pancreatic Cancer. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apc.2019.10.01). The series “Systemic and Targeted Therapies for Pancreas Ductal Adenocarcinoma” was commissioned by the editorial office without any funding or sponsorship. N Ahuja has licensed pancreas cancer biomarkers to Cepheid and has received grant funding from Astex Inc. She has served as a consultant for Ethicon. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913-21. [Crossref] [PubMed]
- Conroy T, Hammel P, Hebbar M, et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N Engl J Med 2018;379:2395-406. [Crossref] [PubMed]
- Wang-Gillam A, Li CP, Bodoky G, et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomised, open-label, phase 3 trial. Lancet 2016;387:545-57. [Crossref] [PubMed]
- Wang-Gillam A, Von Hoff D, Siveke J, et al. Nanoliposomal Irinotecan in the Clinical Practice Guideline for Metastatic Pancreatic Cancer: Applicability to Clinical Situations. J Clin Oncol 2017;35:689-90. [Crossref] [PubMed]
- Biswas S, Rao CM. Epigenetics in cancer: Fundamentals and Beyond. Pharmacol Ther 2017;173:118-34. [Crossref] [PubMed]
- Holliday R. DNA methylation and epigenetic mechanisms. Cell Biophys 1989;15:15-20. [Crossref] [PubMed]
- Azad N, Zahnow CA, Rudin CM, et al. The future of epigenetic therapy in solid tumours--lessons from the past. Nat Rev Clin Oncol 2013;10:256-66. [Crossref] [PubMed]
- Kouzarides T. Chromatin modifications and their function. Cell 2007;128:693-705. [Crossref] [PubMed]
- Schuettengruber B, Chourrout D, Vervoort M, et al. Genome regulation by polycomb and trithorax proteins. Cell 2007;128:735-45. [Crossref] [PubMed]
- Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 2010;11:264-75. [Crossref] [PubMed]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21:381-95. [Crossref] [PubMed]
- Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016;17:284-99. [Crossref] [PubMed]
- Ahuja N, Easwaran H, Baylin SB. Harnessing the potential of epigenetic therapy to target solid tumors. J Clin Invest 2014;124:56-63. [Crossref] [PubMed]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006;6:107-16. [Crossref] [PubMed]
- Dawson MA. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science 2017;355:1147-52. [Crossref] [PubMed]
- Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148-59. [Crossref] [PubMed]
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399-405. [Crossref] [PubMed]
- Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Sato N, Fukushima N, Maitra A, et al. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res 2003;63:3735-42. [PubMed]
- Fukushima N, Sato N, Ueki T, et al. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol 2002;160:1573-81. [Crossref] [PubMed]
- Yi JM, Guzzetta AA, Bailey VJ, et al. Novel methylation biomarker panel for the early detection of pancreatic cancer. Clin Cancer Res 2013;19:6544-55. [Crossref] [PubMed]
- Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov 2012;2:405-13. [Crossref] [PubMed]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042-54. [Crossref] [PubMed]
- Jair KW, Bachman KE, Suzuki H, et al. De novo CpG island methylation in human cancer cells. Cancer Res 2006;66:682-92. [Crossref] [PubMed]
- Kang KA, Piao MJ, Ryu YS, et al. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget 2016;7:40594-620. [Crossref] [PubMed]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597-610. [Crossref] [PubMed]
- Klutstein M, Nejman D, Greenfield R, et al. DNA Methylation in Cancer and Aging. Cancer Res 2016;76:3446-50. [Crossref] [PubMed]
- Li A, Omura N, Hong SM, et al. Pancreatic cancer DNMT1 expression and sensitivity to DNMT1 inhibitors. Cancer Biol Ther 2010;9:321-9. [Crossref] [PubMed]
- Kuroki T, Tajima Y, Kanematsu T. Role of hypermethylation on carcinogenesis in the pancreas. Surg Today 2004;34:981-6. [Crossref] [PubMed]
- Sato N, Goggins M. The role of epigenetic alterations in pancreatic cancer. J Hepatobiliary Pancreat Surg 2006;13:286-95. [Crossref] [PubMed]
- Tan AC, Jimeno A, Lin SH, et al. Characterizing DNA methylation patterns in pancreatic cancer genome. Mol Oncol 2009;3:425-38. [Crossref] [PubMed]
- Schutte M, Hruban RH, Geradts J, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res 1997;57:3126-30. [PubMed]
- Eissa MAL, Lerner L, Abdelfatah E, et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin Epigenetics 2019;11:59. [Crossref] [PubMed]
- Navarro A, Yin P, Monsivais D, et al. Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS One 2012;7:e33284 [Crossref] [PubMed]
- Iguchi E, Safgren SL, Marks DL, et al. Pancreatic Cancer, A Mis-interpreter of the Epigenetic Language. Yale J Biol Med 2016;89:575-90. [PubMed]
- Vincent A, Omura N, Hong SM, et al. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res 2011;17:4341-54. [Crossref] [PubMed]
- Blockus H, Chedotal A. Slit-Robo signaling. Development 2016;143:3037-44. [Crossref] [PubMed]
- Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E, et al. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell 2005;7:39-49. [Crossref] [PubMed]
- Henriksen SD, Madsen PH, Larsen AC, et al. Promoter hypermethylation in plasma-derived cell-free DNA as a prognostic marker for pancreatic adenocarcinoma staging. Int J Cancer 2017;141:2489-97. [Crossref] [PubMed]
- Jäkel C, Bergmann F, Toth R, et al. Genome-wide genetic and epigenetic analyses of pancreatic acinar cell carcinomas reveal aberrations in genome stability. Nat Commun 2017;8:1323. [Crossref] [PubMed]
- Neureiter D, Jager T, Ocker M, et al. Epigenetics and pancreatic cancer: pathophysiology and novel treatment aspects. World J Gastroenterol 2014;20:7830-48. [Crossref] [PubMed]
- Kaminskas E, Farrell AT, Wang YC, et al. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist 2005;10:176-82. [Crossref] [PubMed]
- Santos FP, Kantarjian H, Garcia-Manero G, et al. Decitabine in the treatment of myelodysplastic syndromes. Expert Rev Anticancer Ther 2010;10:9-22. [Crossref] [PubMed]
- Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol 2009;10:223-32. [Crossref] [PubMed]
- Kubasch AS, Platzbecker U. Beyond the Edge of Hypomethylating Agents: Novel Combination Strategies for Older Adults with Advanced MDS and AML. Cancers (Basel) 2018; [Crossref] [PubMed]
- Kumari A, Srinivasan R, Wig JD. Effect of c-MYC and E2F1 gene silencing and of 5-azacytidine treatment on telomerase activity in pancreatic cancer-derived cell lines. Pancreatology 2009;9:360-8. [Crossref] [PubMed]
- Ramachandran K, Miller H, Gordian E, et al. Methylation-mediated silencing of TMS1 in pancreatic cancer and its potential contribution to chemosensitivity. Anticancer Res 2010;30:3919-25. [PubMed]
- Yang H, Lu X, Qian J, et al. Imprinted tumor suppressor gene ARHI induces apoptosis correlated with changes in DNA methylation in pancreatic cancer cells. Mol Med Rep 2010;3:581-7. [PubMed]
- Gupta S, Pramanik D, Mukherjee R, et al. Molecular determinants of retinoic acid sensitivity in pancreatic cancer. Clin Cancer Res 2012;18:280-9. [Crossref] [PubMed]
- Pan FP, Zhou HK, Bu HQ, et al. Emodin enhances the demethylation by 5-Aza-CdR of pancreatic cancer cell tumor-suppressor genes P16, RASSF1A and ppENK. Oncol Rep 2016;35:1941-9. [Crossref] [PubMed]
- Wang X, Wang H, Jiang N, et al. Effect of inhibition of MEK pathway on 5-aza-deoxycytidine-suppressed pancreatic cancer cell proliferation. Genet Mol Res 2013;12:5560-73. [Crossref] [PubMed]
- Missiaglia E, Donadelli M, Palmieri M, et al. Growth delay of human pancreatic cancer cells by methylase inhibitor 5-aza-2'-deoxycytidine treatment is associated with activation of the interferon signalling pathway. Oncogene 2005;24:199-211. [Crossref] [PubMed]
- Thakar M, Hu Y, Morreale M, et al. A novel epigenetic modulating agent sensitizes pancreatic cells to a chemotherapy agent. PLoS One 2018;13:e0199130 [Crossref] [PubMed]
- Epelbaum R, Schaffer M, Vizel B, et al. Curcumin and gemcitabine in patients with advanced pancreatic cancer. Nutr Cancer 2010;62:1137-41. [Crossref] [PubMed]
- Kanai M, Yoshimura K, Asada M, et al. A phase I/II study of gemcitabine-based chemotherapy plus curcumin for patients with gemcitabine-resistant pancreatic cancer. Cancer Chemother Pharmacol 2011;68:157-64. [Crossref] [PubMed]
- Dhillon N, Aggarwal BB, Newman RA, et al. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res 2008;14:4491-9. [Crossref] [PubMed]
- Prébet T, Gore SD, Esterni B, et al. Outcome of high-risk myelodysplastic syndrome after azacitidine treatment failure. J Clin Oncol 2011;29:3322-7. [Crossref] [PubMed]
- Qin T, Castoro R, El Ahdab S, et al. Mechanisms of resistance to decitabine in the myelodysplastic syndrome. PLoS One 2011;6:e23372 [Crossref] [PubMed]
- Stewart DJ, Issa JP, Kurzrock R, et al. Decitabine effect on tumor global DNA methylation and other parameters in a phase I trial in refractory solid tumors and lymphomas. Clin Cancer Res 2009;15:3881-8. [Crossref] [PubMed]
- Tsai HC, Li H, Van Neste L, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 2012;21:430-46. [Crossref] [PubMed]
- Oki Y, Jelinek J, Shen L, et al. Induction of hypomethylation and molecular response after decitabine therapy in patients with chronic myelomonocytic leukemia. Blood 2008;111:2382-4. [Crossref] [PubMed]
- Allis CD, Berger SL, Cote J, et al. New nomenclature for chromatin-modifying enzymes. Cell 2007;131:633-6. [Crossref] [PubMed]
- Bartke T, Kouzarides T. Decoding the chromatin modification landscape. Cell Cycle 2011;10:182. [Crossref] [PubMed]
- de Ruijter AJ, van Gennip AH, Caron HN, et al. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 2003;370:737-49. [Crossref] [PubMed]
- Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J Cell Biochem 2002;87:117-25. [Crossref] [PubMed]
- Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005;435:1262-6. [Crossref] [PubMed]
- Barlési F, Giaccone G, Gallegos-Ruiz MI, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. J Clin Oncol 2007;25:4358-64. [Crossref] [PubMed]
- Park YS, Jin MY, Kim YJ, et al. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann Surg Oncol 2008;15:1968-76. [Crossref] [PubMed]
- Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 2005;37:391-400. [Crossref] [PubMed]
- Watanabe T, Morinaga S, Akaike M, et al. The cellular level of histone H3 lysine 4 dimethylation correlates with response to adjuvant gemcitabine in Japanese pancreatic cancer patients treated with surgery. Eur J Surg Oncol 2012;38:1051-7. [Crossref] [PubMed]
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 2008;647:21-9. [Crossref] [PubMed]
- Agarwal P, Jackson SP. G9a inhibition potentiates the anti-tumour activity of DNA double-strand break inducing agents by impairing DNA repair independent of p53 status. Cancer Lett 2016;380:467-75. [Crossref] [PubMed]
- Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 2002;16:1779-91. [Crossref] [PubMed]
- Pan MR, Hsu MC, Luo CW, et al. The histone methyltransferase G9a as a therapeutic target to override gemcitabine resistance in pancreatic cancer. Oncotarget 2016;7:61136-51. [Crossref] [PubMed]
- Chen YT, Zhu F, Lin WR, et al. The novel EZH2 inhibitor, GSK126, suppresses cell migration and angiogenesis via down-regulating VEGF-A. Cancer Chemother Pharmacol 2016;77:757-65. [Crossref] [PubMed]
- Mathison A, Salmonson A, Missfeldt M, et al. Combined AURKA and H3K9 Methyltransferase Targeting Inhibits Cell Growth By Inducing Mitotic Catastrophe. Mol Cancer Res 2017;15:984-97. [Crossref] [PubMed]
- McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012;492:108-12. [Crossref] [PubMed]
- Ougolkov AV, Bilim VN, Billadeau DD. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin Cancer Res 2008;14:6790-6. [Crossref] [PubMed]
- Hung SW, Mody H, Marrache S, et al. Pharmacological reversal of histone methylation presensitizes pancreatic cancer cells to nucleoside drugs: in vitro optimization and novel nanoparticle delivery studies. PLoS One 2013;8:e71196 [Crossref] [PubMed]
- Avan A, Crea F, Paolicchi E, et al. Molecular mechanisms involved in the synergistic interaction of the EZH2 inhibitor 3-deazaneplanocin A with gemcitabine in pancreatic cancer cells. Mol Cancer Ther 2012;11:1735-46. [Crossref] [PubMed]
- Jiao F, Hu H, Yuan C, et al. Histone deacetylase 3 promotes pancreatic cancer cell proliferation, invasion and increases drug-resistance through histone modification of P27, P53 and Bax. Int J Oncol 2014;45:1523-30. [Crossref] [PubMed]
- Ouaïssi M, Sielezneff I, Silvestre R, et al. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol 2008;15:2318-28. [Crossref] [PubMed]
- Fritsche P, Seidler B, Schuler S, et al. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut 2009;58:1399-409. [Crossref] [PubMed]
- Schüler S, Fritsche P, Diersch S, et al. HDAC2 attenuates TRAIL-induced apoptosis of pancreatic cancer cells. Mol Cancer 2010;9:80. [Crossref] [PubMed]
- von Burstin J, Eser S, Paul MC, et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology 2009;137:361-71, 371.e1-5.
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12-27. [Crossref] [PubMed]
- Moore PS, Barbi S, Donadelli M, et al. Gene expression profiling after treatment with the histone deacetylase inhibitor trichostatin A reveals altered expression of both pro- and anti-apoptotic genes in pancreatic adenocarcinoma cells. Biochim Biophys Acta 2004;1693:167-76. [Crossref] [PubMed]
- Ammerpohl O, Trauzold A, Schniewind B, et al. Complementary effects of HDAC inhibitor 4-PB on gap junction communication and cellular export mechanisms support restoration of chemosensitivity of PDAC cells. Br J Cancer 2007;96:73-81. [Crossref] [PubMed]
- Jung KH, Noh JH, Kim JK, et al. HDAC2 overexpression confers oncogenic potential to human lung cancer cells by deregulating expression of apoptosis and cell cycle proteins. J Cell Biochem 2012;113:2167-77. [Crossref] [PubMed]
- Fortson WS, Kayarthodi S, Fujimura Y, et al. Histone deacetylase inhibitors, valproic acid and trichostatin-A induce apoptosis and affect acetylation status of p53 in ERG-positive prostate cancer cells. Int J Oncol 2011;39:111-9. [PubMed]
- Mann BS, Johnson JR, Cohen MH, et al. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 2007;12:1247-52. [Crossref] [PubMed]
- Coiffier B, Pro B, Prince HM, et al. Romidepsin for the treatment of relapsed/refractory peripheral T-cell lymphoma: pivotal study update demonstrates durable responses. J Hematol Oncol 2014;7:11. [Crossref] [PubMed]
- Yee AJ, Raje NS. Panobinostat and Multiple Myeloma in 2018. Oncologist 2018;23:516-7. [Crossref] [PubMed]
- Jones SF, Bendell JC, Infante JR, et al. A phase I study of panobinostat in combination with gemcitabine in the treatment of solid tumors. Clin Adv Hematol Oncol 2011;9:225-30. [PubMed]
- Chan E, Chiorean EG, O'Dwyer PJ, et al. Phase I/II study of mocetinostat in combination with gemcitabine for patients with advanced pancreatic cancer and other advanced solid tumors. Cancer Chemother Pharmacol 2018;81:355-64. [Crossref] [PubMed]
- Jones SF, Infante JR, Spigel DR, et al. Phase 1 results from a study of romidepsin in combination with gemcitabine in patients with advanced solid tumors. Cancer Invest 2012;30:481-6. [Crossref] [PubMed]
- Lassen U, Molife LR, Sorensen M, et al. A phase I study of the safety and pharmacokinetics of the histone deacetylase inhibitor belinostat administered in combination with carboplatin and/or paclitaxel in patients with solid tumours. Br J Cancer 2010;103:12-7. [Crossref] [PubMed]
- Pili R, Salumbides B, Zhao M, et al. Phase I study of the histone deacetylase inhibitor entinostat in combination with 13-cis retinoic acid in patients with solid tumours. Br J Cancer 2012;106:77-84. [Crossref] [PubMed]
- Iwahashi S, Utsunomiya T, Imura S, et al. Effects of valproic acid in combination with S-1 on advanced pancreatobiliary tract cancers: clinical study phases I/II. Anticancer Res 2014;34:5187-91. [PubMed]
- Chan E, Arlinghaus LR, Cardin DB, et al. Phase I trial of vorinostat added to chemoradiation with capecitabine in pancreatic cancer. Radiother Oncol 2016;119:312-8. [Crossref] [PubMed]
- Richards DA, Boehm KA, Waterhouse DM, et al. Gemcitabine plus CI-994 offers no advantage over gemcitabine alone in the treatment of patients with advanced pancreatic cancer: results of a phase II randomized, double-blind, placebo-controlled, multicenter study. Ann Oncol 2006;17:1096-102. [Crossref] [PubMed]
- Millward M, Price T, Townsend A, et al. Phase 1 clinical trial of the novel proteasome inhibitor marizomib with the histone deacetylase inhibitor vorinostat in patients with melanoma, pancreatic and lung cancer based on in vitro assessments of the combination. Invest New Drugs 2012;30:2303-17. [Crossref] [PubMed]
- Wang H, Cao Q, Dudek AZ. Phase II study of panobinostat and bortezomib in patients with pancreatic cancer progressing on gemcitabine-based therapy. Anticancer Res 2012;32:1027-31. [PubMed]
- Nguyen AH, Elliott IA, Wu N, et al. Histone deacetylase inhibitors provoke a tumor supportive phenotype in pancreatic cancer associated fibroblasts. Oncotarget 2017;8:19074-88. [Crossref] [PubMed]
- Miller KM, Tjeertes JV, Coates J, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010;17:1144-51. [Crossref] [PubMed]
- Chen YJ, Wang WH, Wu WY, et al. Novel histone deacetylase inhibitor AR-42 exhibits antitumor activity in pancreatic cancer cells by affecting multiple biochemical pathways. PLoS One 2017;12:e0183368 [Crossref] [PubMed]
- Golan T, Javle M. DNA Repair Dysfunction in Pancreatic Cancer: A Clinically Relevant Subtype for Drug Development. J Natl Compr Canc Netw 2017;15:1063-9. [Crossref] [PubMed]
- Münster P, Marchion D, Bicaku E, et al. Phase I trial of histone deacetylase inhibition by valproic acid followed by the topoisomerase II inhibitor epirubicin in advanced solid tumors: a clinical and translational study. J Clin Oncol 2007;25:1979-85. [Crossref] [PubMed]
- Postel-Vinay S, Herbschleb K, Massard C, et al. First-in-human phase I study of the bromodomain and extraterminal motif inhibitor BAY 1238097: emerging pharmacokinetic/pharmacodynamic relationship and early termination due to unexpected toxicity. Eur J Cancer 2019;109:103-10. [Crossref] [PubMed]
- Mross K, Dittrich C, Aulitzky WE, et al. A randomised phase II trial of the Polo-like kinase inhibitor BI 2536 in chemo-naive patients with unresectable exocrine adenocarcinoma of the pancreas - a study within the Central European Society Anticancer Drug Research (CESAR) collaborative network. Br J Cancer 2012;107:280-6. [Crossref] [PubMed]
- Koenig A, Linhart T, Schlengemann K, et al. NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology 2010;138:1189-99.e1-2.
- Mees ST, Mardin WA, Sielker S, et al. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol 2009;16:2339-50. [Crossref] [PubMed]
- Pérez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics 2017;12:323-39. [Crossref] [PubMed]
- Sahai V, Redig AJ, Collier KA, et al. Targeting BET bromodomain proteins in solid tumors. Oncotarget 2016;7:53997-4009. [Crossref] [PubMed]
- Noguchi-Yachide T. BET Bromodomain as a Target of Epigenetic Therapy. Chem Pharm Bull (Tokyo) 2016;64:540-7. [Crossref] [PubMed]
- French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol 2012;7:247-65. [Crossref] [PubMed]
- Bauer DE, Mitchell CM, Strait KM, et al. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin Cancer Res 2012;18:5773-9. [Crossref] [PubMed]
- Stathis A, Zucca E, Bekradda M, et al. Clinical Response of Carcinomas Harboring the BRD4-NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer Discov 2016;6:492-500. [Crossref] [PubMed]
- Lewin J, Soria J, Stathis A, et al. Phase Ib trial with birabresib, a small-molecule inhibitor of bromodomain and extraterminal proteins, in patients with selected advanced solid tumors. J Clin Oncol 2018;36:3007-14. [Crossref] [PubMed]
- Hilton J, Cristea MC, Voskoboynik M, et al. Initial results from a phase I/IIa trial evaluating BMS986158, an inhibitor of the bromodomain and extra-terminal (BET) proteins, in patients (pts) with advanced cancer. Ann Oncol 2018;29:viii133-viii148.
- Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146:904-17. [Crossref] [PubMed]
- Grayson AR, Walsh EM, Cameron MJ, et al. MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene 2014;33:1736-42. [Crossref] [PubMed]
- Shi J, Wang Y, Zeng L, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 2014;25:210-25. [Crossref] [PubMed]
- Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067-73. [Crossref] [PubMed]
- Mazur PK, Herner A, Mello SS, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med 2015;21:1163-71. [Crossref] [PubMed]
- Wang YH, Sui XM, Sui YN, et al. BRD4 induces cell migration and invasion in HCC cells through MMP-2 and MMP-9 activation mediated by the Sonic hedgehog signaling pathway. Oncol Lett 2015;10:2227-32. [Crossref] [PubMed]
- Huang Y, Nahar S, Nakagawa A, et al. Regulation of GLI Underlies a Role for BET Bromodomains in Pancreatic Cancer Growth and the Tumor Microenvironment. Clin Cancer Res 2016;22:4259-70. [Crossref] [PubMed]
- Sahai V, Kumar K, Knab LM, et al. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol Cancer Ther 2014;13:1907-17. [Crossref] [PubMed]
- Garcia PL, Miller AL, Kreitzburg KM, et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene 2016;35:833-45. [Crossref] [PubMed]
- Leal AS, Williams CR, Royce DB, et al. Bromodomain inhibitors, JQ1 and I-BET 762, as potential therapies for pancreatic cancer. Cancer Lett 2017;394:76-87. [Crossref] [PubMed]
- Yamamoto K, Tateishi K, Kudo Y, et al. Stromal remodeling by the BET bromodomain inhibitor JQ1 suppresses the progression of human pancreatic cancer. Oncotarget 2016;7:61469-84. [Crossref] [PubMed]
- Bhadury J, Nilsson LM, Muralidharan SV, et al. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc Natl Acad Sci U S A 2014;111:E2721-30. [Crossref] [PubMed]
- Fiskus W, Sharma S, Qi J, et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther 2014;13:1142-54. [Crossref] [PubMed]
- Cote GA, Gore AJ, McElyea SD, et al. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am J Gastroenterol 2014;109:1942-52. [Crossref] [PubMed]
- Habbe N, Koorstra JB, Mendell JT, et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther 2009;8:340-6. [Crossref] [PubMed]
- Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A 2007;104:16170-5. [Crossref] [PubMed]
- Xue Y, Abou Tayoun AN, Abo KM, et al. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet 2013;206:217-21. [Crossref] [PubMed]
- Yu J, Li A, Hong SM, et al. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res 2012;18:981-92. [Crossref] [PubMed]
- du Rieu MC, Torrisani J, Selves J, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem 2010;56:603-12. [Crossref] [PubMed]
- Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007;297:1901-8. [Crossref] [PubMed]
- Vila-Navarro E, Vila-Casadesus M, Moreira L, et al. MicroRNAs for Detection of Pancreatic Neoplasia: Biomarker Discovery by Next-generation Sequencing and Validation in 2 Independent Cohorts. Ann Surg 2017;265:1226-34. [Crossref] [PubMed]
- Ma MZ, Kong X, Weng MZ, et al. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res 2013;32:71. [Crossref] [PubMed]
- Wan C, Shen Y, Yang T, et al. Diagnostic value of microRNA for pancreatic cancer: a meta-analysis. Arch Med Sci 2012;8:749-55. [Crossref] [PubMed]
- Kong X, Du Y, Wang G, et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci 2011;56:602-9. [Crossref] [PubMed]
- Ikenaga N, Ohuchida K, Mizumoto K, et al. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol 2010;17:3120-8. [Crossref] [PubMed]
- Zhou L, Zhang WG, Wang DS, et al. MicroRNA-183 is involved in cell proliferation, survival and poor prognosis in pancreatic ductal adenocarcinoma by regulating Bmi-1. Oncol Rep 2014;32:1734-40. [Crossref] [PubMed]
- Yao J, Li Z, Wang X, et al. MiR-125a regulates chemo-sensitivity to gemcitabine in human pancreatic cancer cells through targeting A20. Acta Biochim Biophys Sin (Shanghai) 2016;48:202-8. [Crossref] [PubMed]
- Hwang JH, Voortman J, Giovannetti E, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One 2010;5:e10630 [Crossref] [PubMed]
- Moriyama T, Ohuchida K, Mizumoto K, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther 2009;8:1067-74. [Crossref] [PubMed]
- Cai B, An Y, Lv N, et al. miRNA-181b increases the sensitivity of pancreatic ductal adenocarcinoma cells to gemcitabine in vitro and in nude mice by targeting BCL-2. Oncol Rep 2013;29:1769-76. [Crossref] [PubMed]
- Wang P, Zhang J, Zhang L, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology 2013;145:1133-43.e12. [Crossref] [PubMed]
- Lomberk GA, Iovanna J, Urrutia R. The promise of epigenomic therapeutics in pancreatic cancer. Epigenomics 2016;8:831-42. [Crossref] [PubMed]
- Zheng J, Huang X, Tan W, et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet 2016;48:747-57. [Crossref] [PubMed]
- Ma C, Nong K, Zhu H, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol 2014;35:9163-9. [Crossref] [PubMed]
- Ma L, Tian X, Wang F, et al. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther 2016;17:1051-61. [Crossref] [PubMed]
- Xie Z, Chen X, Li J, et al. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 2016;7:25408-19. [PubMed]
- Liu C, Wang J, Yuan X, et al. Long noncoding RNA uc.345 promotes tumorigenesis of pancreatic cancer by upregulation of hnRNPL expression. Oncotarget 2016;7:71556-66. [PubMed]
- Li L, Zhang GQ, Chen H, et al. Plasma and tumor levels of Linc-pint are diagnostic and prognostic biomarkers for pancreatic cancer. Oncotarget 2016;7:71773-81. [PubMed]
- Pai P, Rachagani S, Are C, et al. Prospects of miRNA-based therapy for pancreatic cancer. Curr Drug Targets 2013;14:1101-9. [Crossref] [PubMed]
- Xia J, Duan Q, Ahmad A, et al. Genistein inhibits cell growth and induces apoptosis through up-regulation of miR-34a in pancreatic cancer cells. Curr Drug Targets 2012;13:1750-6. [Crossref] [PubMed]
- Pramanik D, Campbell NR, Karikari C, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther 2011;10:1470-80. [Crossref] [PubMed]
- Mittal A, Chitkara D, Behrman SW, et al. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials 2014;35:7077-87. [Crossref] [PubMed]
- Zhao Y, Zhao L, Ischenko I, et al. Antisense inhibition of microRNA-21 and microRNA-221 in tumor-initiating stem-like cells modulates tumorigenesis, metastasis, and chemotherapy resistance in pancreatic cancer. Target Oncol 2015;10:535-48. [Crossref] [PubMed]
- Park JK, Henry JC, Jiang J, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun 2011;406:518-23. [Crossref] [PubMed]
- El-Rayes BF, Philip PA, Sarkar FH, et al. A phase II study of isoflavones, erlotinib, and gemcitabine in advanced pancreatic cancer. Invest New Drugs 2011;29:694-9. [Crossref] [PubMed]
- Sarkar S, Dubaybo H, Ali S, et al. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res 2013;3:465-77. [PubMed]
- Hanna N, Ohana P, Konikoff FM, et al. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther 2012;19:374-81. [Crossref] [PubMed]
- Brahmer JR, Drake CG, Wollner I, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol 2010;28:3167-75. [Crossref] [PubMed]
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. [Crossref] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Lutz ER, Wu AA, Bigelow E, et al. Immunotherapy converts nonimmunogenic pancreatic tumors into immunogenic foci of immune regulation. Cancer Immunol Res 2014;2:616-31. [Crossref] [PubMed]
- Soares KC, Rucki AA, Wu AA, et al. PD-1/PD-L1 blockade together with vaccine therapy facilitates effector T-cell infiltration into pancreatic tumors. J Immunother 2015;38:1-11. [Crossref] [PubMed]
- Aspeslagh S, Morel D, Soria JC, et al. Epigenetic modifiers as new immunomodulatory therapies in solid tumours. Ann Oncol 2018;29:812-24. [Crossref] [PubMed]
- Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015;162:974-86. [Crossref] [PubMed]
- Wrangle J, Wang W, Koch A, et al. Alterations of immune response of Non-Small Cell Lung Cancer with Azacytidine. Oncotarget 2013;4:2067-79. [Crossref] [PubMed]
- Roulois D, Loo Yau H, Singhania R, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015;162:961-73. [Crossref] [PubMed]
- Cycon KA, Mulvaney K, Rimsza LM, et al. Histone deacetylase inhibitors activate CIITA and MHC class II antigen expression in diffuse large B-cell lymphoma. Immunology 2013;140:259-72. [Crossref] [PubMed]
- Zhu S, Denman CJ, Cobanoglu ZS, et al. The narrow-spectrum HDAC inhibitor entinostat enhances NKG2D expression without NK cell toxicity, leading to enhanced recognition of cancer cells. Pharm Res 2015;32:779-92. [Crossref] [PubMed]
- Zhu H, Bengsch F, Svoronos N, et al. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep 2016;16:2829-37. [Crossref] [PubMed]
- Muralidharan SV, Einarsdottir BO, Bhadury J, et al. BET bromodomain inhibitors synergize with ATR inhibitors in melanoma. Cell Death Dis 2017;8:e2982 [Crossref] [PubMed]
- Peng D, Kryczek I, Nagarsheth N, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015;527:249-53. [Crossref] [PubMed]
- Shen L, Ciesielski M, Ramakrishnan S, et al. Class I histone deacetylase inhibitor entinostat suppresses regulatory T cells and enhances immunotherapies in renal and prostate cancer models. PLoS One 2012;7:e30815 [Crossref] [PubMed]
- Kim K, Skora AD, Li Z, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc Natl Acad Sci U S A 2014;111:11774-9. [Crossref] [PubMed]
- Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011;1:598-607. [Crossref] [PubMed]
- Gandhi L, Janne PA, Opyrchal M, et al. Efficacy an safety of entinostat (ENT) and pembrolizumab (PEMBRO) in patients with non-small cell lung cancer (NSCLC) previously treated with anti-PD-(L)1 therapy J Clin Oncol 2018;36:9036. [Crossref]
- Li H, Chiappinelli KB, Guzzetta AA, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014;5:587-98. [Crossref] [PubMed]
- Clark CE, Hingorani SR, Mick R, et al. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res 2007;67:9518-27. [Crossref] [PubMed]
- Skelton RA, Javed A, Zheng L, et al. Overcoming the resistance of pancreatic cancer to immune checkpoint inhibitors. J Surg Oncol 2017;116:55-62. [Crossref] [PubMed]
- von Bernstorff W, Voss M, Freichel S, et al. Systemic and local immunosuppression in pancreatic cancer patients. Clin Cancer Res 2001;7:925s-32s. [PubMed]
- Shakya R, Gonda T, Quante M, et al. Hypomethylating therapy in an aggressive stroma-rich model of pancreatic carcinoma. Cancer Res 2013;73:885-96. [Crossref] [PubMed]
- Lu C, Paschall AV, Shi H, et al. The MLL1-H3K4me3 Axis-Mediated PD-L1 Expression and Pancreatic Cancer Immune Evasion. J Natl Cancer Inst 2017; [Crossref] [PubMed]
- Christmas BJ, Rafie CI, Hopkins AC, et al. Entinostat Converts Immune-Resistant Breast and Pancreatic Cancers into Checkpoint-Responsive Tumors by Reprogramming Tumor-Infiltrating MDSCs. Cancer Immunol Res 2018;6:1561-77. [Crossref] [PubMed]
- Baretti M, Durham JN, Walker R, et al. Entinostat in combination with nivolumab for patients with advanced cholangiocarcinoma and pancreatic adenocarcinoma. Journal of Clinical Oncology 2018;36:TPS4151 [Crossref]
- McCleary-Wheeler AL, Lomberk GA, Weiss FU, et al. Insights into the epigenetic mechanisms controlling pancreatic carcinogenesis. Cancer Lett 2013;328:212-21. [Crossref] [PubMed]
- Hessmann E, Johnsen SA, Siveke JT, et al. Epigenetic treatment of pancreatic cancer: is there a therapeutic perspective on the horizon? Gut 2017;66:168-79. [Crossref] [PubMed]
Cite this article as: Baretti M, Ahuja N, Azad NS. Targeting the epigenome of pancreatic cancer for therapy: challenges and opportunities. Ann Pancreat Cancer 2019;2:18.


