
Targeting homologous recombination addicted tumors: challenges and opportunities
Background on the discovery, cloning, and importance of genome maintenance and DNA repair genes
Linking BRCA1/2 to cancer predisposition: Let the games begin
In the early 1990s, BRCA1 was mapped to chromosome 17 (1), and thus, initiated the international effort to link half the families from a consortium with breast and ovarian cancers (an inherited predisposition) to this loci (2). This investigation, amongst others, pioneered the efforts to discover inherited germline mutations in genes that could explain the reason why some families had a strong predisposition to certain tumors. For instance, it is well established that individuals who carry a germline heterozygous BRCA1 (or BRCA2) mutation can have a lifetime risk of ~85% of developing breast and ovarian cancers (3), and a higher incidence of developing pancreatic and prostate cancers than the general population. These genetic linkage studies were not only enlightening for genetic testing and identifying cancer predisposition syndromes, which have highlighted the importance of DNA repair for cancer prevention (4), but also opened the field to a new understanding about the intersection between a defective DDR pathway and tumorigenesis. Finally, the culmination of this work plus the sequencing of thousands of cancer genomes have underscored the importance of the DDR pathway. For instance, to date the clinical concept of personalizing therapy for the treatment of DDR defective tumors is very much in vogue.
Although BRCA1/2 have been implicated in a number of cellular functions, the fact that these genes are mutated with a high frequency in tumors and have a well described genome maintenance and repair mechanism, has led the field to focus on its role in tumorigenesis (5). Specifically, BRCA1/2 have been established to play a key role in collapsed replication forks and double strand DNA break (DSBs) via homologous recombination (HR), where the damaged site in DNA sequenced is repaired in a conventional manner. In a deficient BRCA1/2 setting, cells use a faulty, alternative repair mechanism, and this is believed to facilitate genetic instability and tumorigenesis (6).
From genes to an homologous recombination deficiency (HRD) signature/scar: are all DDR genes mutated equally?
The discovery of DDR genes linked to cancer predisposition and tumorigenesis has forced NGS panels to expand personalized approaches to think beyond BRCAness (i.e., BRCA1/2 genes). However, there are limitations to just trying to expand the panel of DDR genes. First, it is unclear whether low frequency mutated DDR genes (or even variants) are truly driver alterations of tumorigenesis. Unfortunately, in many instances, including BRCA1/2 mutated tumors, the frequency of mutations found in a specific tumor type, may be very low compared to more garden variety cancer driving genes (e.g., Kras or TP53), making it hard to decipher whether these events are frequently selected for in a given patient cohort. Based on the central dogma of conventional cancer genetics, one must have a mutation frequency in a tumor type which is greater than expected in a cohort of healthy controls (7). Other layers of complexity include whether these DDR-related genes will have the same Achilles Heel (also known as synthetic lethality) that established DDR genes such as BRCA1/2 have and whether these genes fit the classical tumor suppressor rules of needing to have loss of the second allele in the tumor (e.g., Loss of Heterozygosity, LOH) (7). Therefore, in many DDR genes, it is unclear whether these DDR defective genes have predictive therapeutic value. Based on these concerns, many investigators have attempted to design assays that detect a molecular signature that could identify tumors with a defective DDR pathway (i.e., HRD, see below section).
Specifically, a number of additional genes have been implicated beyond BRCA1/2 genes in the HR pathway. It should be noted that these mutations can be categorized and identified as either germline (i.e., constitutional DNA, identified in the normal blood or presumable inherited) or somatic (i.e., found in the tumor DNA, but not in the germline DNA) in nature. As the field rapidly moves into the next versions of NGS, many of these early concepts or the central dogma of cancer genetics sometimes gets lost in the commercialization of personalized medicine. The gold standard for identifying an actionable tumor suppressor or genome maintenance gene mutation, in this instance would be a germline mutation (i.e., that causes an amino acid change) combined with a second, somatic hit mutation in the other allele, typically via an LOH event. Additional considerations include identifying whether a somatic change is authentic based on the availability to obtain pure DNA from the tumor (without contaminating normal cells that can confound sequencing results) and matched normal tissue in order to compare with tumor sequencing results. An additional level of complexity on determining whether these HR-gene mutations are authentic, is the fact that many are considered Variants of Unknown Significance (VUSs). Many VUSs have been uncharacterized and even with the confirmation of a potential second allele hit, the functional significance of these VUSs may either be linked to a non-driving Single Nucleotide Polymorphism (SNP) or a more prevalent previously described mutation from a patient cohort.
With that written, still a number of DDR-HR related genes have been well described and established. In a recent publication focused on Homologous Repair-DDR genes looking at over 52,000 tumors of different origins (e.g., biliary tract, GI stromal, bladder, etc.) were analyzed to identify pathogenic mutations in the HR-DDR genes including BRCA1/2, BRIP1, RAD50, RAD51, RAD51B, ATM, ATRX, BARD1, CHEK1/2, FANCA/C/D2/E/F/G/L, MRE11A, NBN, and PALB2 (8). In this study, they found that the overall frequency of these mutations were over 17% across 21 tumor lineages. Other studies have focused on specific lineages, such as a recent study by Pancreatic Cancer Action Network with the Know Your Tumor program finding that sequencing data from a cohort of over 600 pancreatic cancers with attempted profiling across 44 states showed a frequency of over 8% mutations in DDR genes (9). Although these data do not account for the possibility of the complete loss of the gene, they do support, similar to other large sequencing studies (10,11), that the DDR pathway is critically important in the tumorigenesis process for a number of tumor systems. More recently, Jonsson et al screened over 17,000 tumor profiles across 55 different types of tumors and found the prevalence of 2.7% for BRCA1/2 pathologic germline variants (12). An additional small percentage had somatic mutations. Combing both germline pathogenic and somatic BRCA1/2 mutations accounted for 4.9% and covered at least 38 cancer types of which the majority were pancreatic, breast, ovary, and prostate cancers (12). Interestingly, only 61% of all BRCA1/2 carriers with various cancers harbored a somatic LOH hit in the wild-type BRCA1/2 allele, this number was significantly enriched over the background rate of LOH in tumors with non-pathogenic variants (20%) (12). These data support the notion that in some instances cancers arise in a setting of BRCA1/2 germline mutations independent of complete loss of BRCAness. Mouse modeling of these gene defects support and also refute some of these findings for the development of cancer. For instance, Venkitaraman and colleagues (13) generated a murine model of familial pancreatic cancer (driven by a KRAS G12D mutation) and found that germline heterozygosity for a functional BRCA2 truncation induced pancreatic ductal adenocarcinomas (PDACs). Not following the two-hit paradigm, tumor cells from these animals did not lose a second BRCA2 allele. Complementary to this work, in three out of four PDACs from patients who inherited a classic BRCA2999del5 mutation, did not obtain a LOH hit in the second allele (11). A more recent publication demonstrated two out of three PDX models from three glBRCA PDAC patients, obtained LOH in second allele (14). It may depend when the model systems are developed in relation to the progress of the patient’s clinical course, since we have shown that over the course of the disease the patients can develop therapeutic resistance, thus changing these genomic events (15). Taken together, these studies: (I) point towards the need of a more comprehensive molecular signature for BRCAness (i.e., HRD scoring); (II) an understanding of how germline and somatic BRCA1/2 mutations interact with different tumor systems and lineages; and (III) a caution to the field about completely relying both on the central dogma of tumor suppressors in regards to genome maintenance genes and germline testing.
Next generation germline/patient testing: the search for a comprehensive molecular signature
Initially, identifying patients with a deficiency in the DDR pathway was solely based on Sanger Sequencing. The use of bench techniques to study DNA damage were, and still are, being explored as a simple test to find a defect in a pathway, without depicting the exact genetic lesion. For instance, by running a FAND2 monoubiquitination assay (Western or immunofluorescence), a molecular lab could determine whether a mutation occurred in the relevant Fanconi Anemia pathway, without knowing the exact gene that is mutated or loss in a cancer (16,17). The rapid development of molecular and –omic signatures have been established since the advent of sequencing, microarray, and proteomic technologies have gotten more advanced and facile. The repertoire and diversity of mutational signatures span from “one-off” publications (i.e., no follow-up or validating study) depicting the prognostic value of a sampling of gene expression levels for the prognostic value and predictive value of a specific therapy. These studies appeared prevalent in the 1990s and 2000s, as the ability to validate a specific focused gene signature in independent cohorts were rare, since large-scale, multi-institutional registry and biobanking strategies were inefficient and rare. More recently, the availability of well-annotated samples, advanced technologies, and rigorously validated publicly available datasets has provided researchers with a reality check on their favorite or novel multi-panel gene signature (18) [for a recent comprehensive review see (19)].
As expected, due to the volume of cases and the amount of available resources (both clinical specimens and funding) the breast and ovarian cancer fields have led the way in developing a molecular signature for a HRD-score (20). Previous scores were solely based on: (I) allelic imbalance in regards to telomeres (21), (II) LOH (22), and (III) large scale genomic instability (23). Over the last few years, others have combined a combination of these markers with advanced technologies and insights to generate HRD scores with the ultimate goal to find a reliable assay that can determine whether a tumor genome is HRD, or even how immune-active these tumors may be (24,25), for the use as a predictive biomarker (14,20,26,27).
A therapeutic opportunity: an example of translating the science to the clinic
As mentioned previously there are differing and overlapping definitions of HRD in cancer. Furthermore, there may be subtle but clinically meaningful differences of HRD signatures per cancer subtype (19). A genomic signature that defines HRD in ovarian cancer may not have the same clinical relevance in PDAC. For example, the Myriad HRD score, was not significantly associated with a higher response rate or prolonged survival in patients treated with FOLFIRINOX in a small retrospective study (28) (see Table 1 list of HRD signatures). The actual prevalence of HRD in PDAC is estimated to be around 10–12% based on whole genome sequencing data (25,30). However, additional efforts to further define subgroups of patients with platinum sensitivity are still evolving, and may expand this subgroup further. Based on the definition of HRD by genomic alterations in DDR pathways (identified from exome sequencing NGS), Pishvaian and colleagues demonstrated a superior overall survival benefit when these patients were exposed to platinum-based chemotherapy. In this study, the percentage of patients demonstrating genomic alterations in DDR pathways was approximately 16% (32).
Table 1
| Signature/algorithm name | Description & features | Reference |
|---|---|---|
| Myriad’s MyChoice HRD | Tested in ovarian tumors and 57 cell lines (breast and pancreatic) | (22) |
| Association between homologous recombination defects and genomic patterns of loss of heterozygosity (LOH) | ||
| The HRD score appears capable of detecting homologous recombination defects regardless of etiology or mechanism | ||
| Single Base Substitution Signatures (SBS3) | Analysis of 7,042 cancers lead to more than 20 distinct mutational signatures | (18) |
| SBS3 signature observed in breast, ovarian and pancreatic tumors showed association with BRA1/2 mutations | ||
| SBS3 is characterized by large deletions (up to 50 bp) with overlapping microhomology at breakpoint junctions | ||
| HRD gene signature | Based on transcriptional profiling approach to systematically identify common molecular changes associated with defective HR repair | (29) |
| Tested on isogenic cell lines established from MCF-10A cells, an immortal human mammary epithelial cell line of nonmalignant origin, with induced deficiency individually in HR repair genes: BRCA1, RAD51 and BRIT1 and other | ||
| HRD gene signature allows interrogation of the status of HR repair by simultaneously considering hundreds of genes and thereby allows identification of HR deficiency in a given cellular state independent of underlying mechanism | ||
| Waddell structural variations load subtyping | Based on whole genome sequencing (WGS) and copy number variation (CNV) analysis of 100 pancreatic ductal adenocarcinomas (PDAC) tumors | (30) |
| Patterns of chromosomal structural variations/rearrangements classified PDACs into 4 subtypes with potential clinical utility: stable, locally rearranged, scattered and unstable | ||
| Genomic instability co-segregated with inactivation of DNA maintenance genes (BRCA1, BRCA2 or PALB2) and a mutational signature of DNA damage repair deficiency | ||
| Double strand break repair signature (DSBR) | Based on whole genome & RNA sequencing on 160 PDAC cases from 154 patients in the discovery cohort and WGS of 95 samples in the replication cohort | (25) |
| Analyses of mutational signatures based on Alexandrov approach identified 4 PDAC principal subtypes: (I) an age-related group dominated by signatures 1 and 5, (II) a double-strand break repair (DSBR) group characterized by signature 3, attributed to deficiencies in homologous recombination repair (HRR) of double-strand breaks; (III) a mismatch repair (MMR) group characterized by signatures 6, 20, and 26, attributed to defects in DNA MMR; and (4) a group characterized by signature 8, of unknown etiology | ||
| DSBR &MMR subtypes were associated with increased expression of antitumor immunity, including activation of CD8-positive T lymphocytes and overexpression of regulatory molecules (CTL4), corresponding to higher frequency of somatic mutations and tumor-specific neoantigens | ||
| HRDetect score | Based on lasso logistic regression model to identify six distinguishing mutational signatures predictive of BRCA1/BRCA2 deficiency | (31) |
| Tested in 560 individuals with breast cancer and validated on independent cohorts of breast, ovarian and pancreatic cancers | ||
| Shows high sensitivity (98.7%) in identification of BRCA1/2 deficient tumors |
Are all DNA damaging agents equally efficacious? And at what time point should these therapies be administered?
It is important to differentiate between the different mechanisms of actions of the chemotherapeutic agents versus PARP inhibitors and additional, emerging targeted DDR drugs in development. These considerations may have a profound clinical impact, since a DDR-deficient tumor may show sensitivity to a DDR related chemotherapy, but not to a specific targeted DDR drug in development (e.g., PARP inhibitors and ATR inhibitors).
The platinum salts (carboplatinum, cisplatin and oxaliplatin), generate covalent cross-links between DNA bases from DNA-damage-inducing chemotherapies. The cytotoxic effects are determined by the relative amount and specific structure of DNA adducts (33). Alkylating agents (e.g., temozolomide) modify DNA bases. Electrophilic alkyl groups covalently bind to cellular nucleophilic sites, including bases in DNA, these interactions are responsible for cytotoxicity (34). Topoisomerases are essential for all organisms as they prevent DNA and RNA entanglements and resolve DNA supercoiling during replication and transcription. Inhibitors of topoisomerase 1 (camptothecin, topotecan and irinotecan) and topoisomerase 2 (etoposide and doxorubicin) generate TOP-DNA adducts and DNA-strand breaks. These drugs generate non-productive TOP-DNA cleavage complexes before re-ligation occurs (34). There are clear similarities and differences between the DNA-damage-inducing chemotherapies, irinotecan and platinum agents are standard of care treatments in PDAC and therefore the most explored in this setting.
A more specific approach to targeting the DDR pathway includes biological therapeutics specifically Poly (ADP-Ribose) Polymerase (PARP) inhibitors for tumors with defects in DNA repair. Tumors with compromised ability to repair double-strand breaks (DSB) by HR, are highly sensitive to blockage of the repair of DNA single-strand breaks (SSB), via the specific and targeted inhibition of PARP. PARP-inhibition causes failure of the repair SSB. This SSB encountered by the replication fork will cause stalling of the fork and therefore may result in fork collapse or the formation of DSB. In the absence of HR functional proteins (e.g., BRCA 1/2), the replication fork cannot be restarted and collapses, causing chromatid breaks. Additional PARP inhibition mechanisms include the “trapping” of PARP-1 protein on the site of DNA damage. This also may interfere with replication fork progression. This approach has demonstrated wide applicability in BRCA associated ovarian, breast, prostate and pancreatic cancer. Furthermore, initial efficacy has also been seen in the treatment of sporadic cancers with additional HR pathway impairments (35).
As mentioned, the most well described HRD biomarker in PDAC is germline BRCA1/2 mutations. The global prevalence of germline BRCA1/2 is around 7% (36). This subgroup of patients have shown a superior overall survival (OS) when treated with platinum based chemotherapy in retrospective studies (37). However, the toxicity profile of platinum treatment including the accumulating neuropathy and hematological toxicity is well described and needs to be considered here (38). PDAC associated with a germline BRCA1/2 mutation demonstrate efficacy to platinum treatment. However, the side effects are debilitating and dose reductions or cessations are usually mandatory, thus limiting the profound therapeutic usefulness in BRCA-associated cancers. Therefore, additional maintenance strategies have been explored. The aim of a maintenance treatment is to provide an alternative treatment approach without compromising the patient’s quality of life. The clinical trial design in maintenance studies, include comparison of drugs in the maintenance setting that have a potentially superior therapeutic window. For instance, the aim of the POLO study: Olaparib as Switch Maintenance Therapy after Response to platinum-based treatment of metastatic germline BRCA-mutant (gBRCAm) pancreatic cancer (36). Patients had to have received a minimum of 16 weeks platinum-based first line chemotherapy, and they had to demonstrate SD or PR or CR in order to be eligible for the clinical trial. Identified patients were randomized in a 2:1 ratio olaparib 300 mg twice daily or placebo. The primary endpoint PFS was 7.4 months on olaparib versus 3.8 months in the placebo arm, HR 0.53 (95% CI: 0.35–0.82; P=0.0038). Interim OS data (at 46% maturity) showed no difference between arms. Final OS results will be evaluated at 69% data maturity. No statistical differences were noted in quality of life measurements between the olaparib versus placebo arm. Olaparib-arm patients were more likely to achieve a response to treatment or maintain disease control; responses were durable lasting a median of over 2 years. Of note, this strategic approach of first-line platinum-based chemotherapy followed by maintenance PARP inhibitor together has an extended PFS benefit to patients with a germline BRCA1/2 mutations and metastatic disease. This study is the first Phase III trial to validate a targeted treatment in a biomarker-selected population of pancreatic cancer patients, highlighting the importance of gBRCAm testing in this setting.
Concluding thoughts and future directions
Clearly both the medical oncology community and our patients diagnosed with cancer are eager to launch into the arena of personalized/precision oncology. In this era of facile genomic sequencing of tumor genomes, identifying defects in DDR genes or finding a relevant HRD score has provided a shiny glimmer of hope. The concept and the success of synthetic lethality for these tumors are real, as clinical trial data keeps emerging that supports this therapeutic strategy in numerous oncologic settings, including maintenance therapy. Simply put, this work is very promising and exciting for a disease like PDAC, where only 9% of patients live 5 years; we may offer a significant subset of patients an enhanced quality of life and overall survival.
Still there are numerous questions and challenges to be worked out as we launch into this next generation of personalized medicine (Table 2). As outlined above, optimizing a rapid screening platform against pure tumor cells will be critical as we attempt to take advantage of a therapeutic window. However, based on recent data, the central dogma concept that two hits in a tumor suppressor or genome maintenance gene may have to be reconsidered in future and ongoing clinical trials in regards to selection criteria and retrospective analyses. Additional ramifications for these findings may not only affect the predictive value of identifying these mutations, but also have diagnostic implications for family members of patients who harbor the same germline mutations. Finally, additional genetic and beyond genomic alterations (e.g., the immune system and post-transcriptional gene regulation) in these tumor related to the DNA repair pathway are still being uncovered (25,39). These molecular lesions and gene regulatory mechanisms within tumor cells may provide novel targets, biomarkers, and important insights into innate and acquired resistance mechanisms for the above described therapies.
Table 2
| Key scientific findings | Clinical implications | Challenges |
|---|---|---|
| Link of BRCA1/2 mutations to cancer | Prognostic and predictive biomarker value | Determining the optimized therapeutic regimen; a deep understanding of the genetics (LOH, haploinsufficiency, etc.) |
| HRD score | Predictive biomarker value | Determining the correct therapy; pure tumor tissue access/evaluation; validating aspects of the score; making facile and economical |
| POLO study | Maintenance therapy for PDAC patients | Identifying patients upfront for maximum benefits |
| Link of DNA repair genes to cancer | Capturing a greater cohort of patients for target therapies | Validating that these genes have the same predictive value as BRCA1/2; determine the significance of VUS or mutations with low frequency within a tumor system |
| Resistance occurs with DNA damaging therapies | Patients recur | Need to better understand genetic and non-genetic mechanisms to overcome resistance |
DDR, DNA damage repair; LOH, loss of heterozygosity; HRD, homologous recombination deficiency; PDAC, pancreatic ductal adenocarcinomas; VUS, variants of unknown significance.
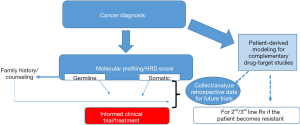
Certainly, a key factor in moving this field forward will be the prospective clinical and molecular analyses of ongoing clinical trials (Figure 1). The recent advances in the development and use of patient derived models of cancer (14,40,41) will provide an invaluable resource to study drug-target (gene) relationships, understanding resistance mechanisms, and may also complement personalized medicine approaches (Figure 1, Table 1). Taken together, we are optimistic that validated multiplexed next generation platforms (that include patient derived models) (Figure 1) that can reliably identify DDR-deficient tumors along with optimized, targeted therapeutic strategies will be a game changer for many lethal cancers (e.g., PDAC) with a DDR-defect.
Acknowledgments
Funding: JRB is supported by NIH-NCI R01 CA212600, the National Cancer Institute of the National Institutes of Health under Award Number P30CA056036 SKCC Core Grant (TJU). 2015 Pancreatic Cancer Action Network American Association for Cancer Research Acceleration Network Grant (15-90-25-BROD). Also the generous support of the W. Kim Foster Pancreatic Cancer Research Endowment. TG is supported by ICRF 2018-2019, MOST-DFZK 2019-2020, Investigator. Initiative Grant Astra Zeneca 2016-2019, Investigator. Initiative Grant Merck MSD 2017-2020, and Soyka Pancreatic Cancer Fund.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apc.2020.03.02). JRB reports personal fees from Perthera, during the conduct of the study. TG: Receipt of grants/research supports: Astra Zeneca and MSD Merck. Receipt of consultation fees: Abbvie, Astra Zeneca, Teva. Bayer and MSD Merck. Receipt of speakers bureau: Abbvie and Bioline. Travel: Astra Zeneca and MSD Merck. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 1990;250:1684-9. [Crossref] [PubMed]
- Easton DF, Bishop DT, Ford D, et al. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The Breast Cancer Linkage Consortium. Am J Hum Genet 1993;52:678-701. [PubMed]
- Wooster R, Weber BL. Breast and ovarian cancer. N Engl J Med 2003;348:2339-47. [Crossref] [PubMed]
- Knoch J, Kamenisch Y, Kubisch C, et al. Rare hereditary diseases with defects in DNA-repair. Eur J Dermatol 2012;22:443-55. [Crossref] [PubMed]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 2006;25:5864-74. [Crossref] [PubMed]
- Tutt A, Bertwistle D, Valentine J, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J 2001;20:4704-16. [Crossref] [PubMed]
- Winter JM, Brody JR, Kern SE. Multiple-criterion evaluation of reported mutations: a proposed scoring system for the intragenic somatic mutation literature. Cancer Biol Ther 2006;5:360-70. [Crossref] [PubMed]
- Heeke AL, Pishvaian MJ, Lynce F, et al. Prevalence of Homologous Recombination-Related Gene Mutations Across Multiple Cancer Types. JCO Precis Oncol 2018;2018: [Crossref] [PubMed]
- Pishvaian MJ, Bender RJ, Halverson D, et al. Molecular Profiling of Patients with Pancreatic Cancer: Initial Results from the Know Your Tumor Initiative. Clin Cancer Res 2018;24:5018-27. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Grant RC, Selander I, Connor AA, et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology 2015;148:556-64. [Crossref] [PubMed]
- Jonsson P, Bandlamudi C, Cheng ML, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature 2019;571:576-9. [Crossref] [PubMed]
- Skoulidis F, Cassidy LD, Pisupati V, et al. Germline Brca2 heterozygosity promotes Kras(G12D) -driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell 2010;18:499-509. [Crossref] [PubMed]
- Golan T, Stossel C, Atias D, et al. Recapitulating the clinical scenario of BRCA-associated pancreatic cancer in pre-clinical models. Int J Cancer 2018;143:179-83. [Crossref] [PubMed]
- Pishvaian MJ, Biankin AV, Bailey P, et al. BRCA2 secondary mutation-mediated resistance to platinum and PARP inhibitor-based therapy in pancreatic cancer. Br J Cancer 2017;116:1021-6. [Crossref] [PubMed]
- van der Heijden MS, Brody JR, Gallmeier E, et al. Functional defects in the fanconi anemia pathway in pancreatic cancer cells. Am J Pathol 2004;165:651-7. [Crossref] [PubMed]
- Van Der Heijden MS, Brody JR, Kern SE. Functional screen of the fanconi anemia pathway in cancer cells by Fancd2 immunoblot. Cancer Biol Ther 2004;3:534-7. [Crossref] [PubMed]
- Petljak M, Alexandrov LB. Understanding mutagenesis through delineation of mutational signatures in human cancer. Carcinogenesis 2016;37:531-40. [Crossref] [PubMed]
- Van Hoeck A, Tjoonk NH, van Boxtel R, et al. Portrait of a cancer: mutational signature analyses for cancer diagnostics. BMC Cancer 2019;19:457. [Crossref] [PubMed]
- Telli ML, Timms KM, Reid J, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res 2016;22:3764-73. [Crossref] [PubMed]
- Birkbak NJ, Wang ZC, Kim JY, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012;2:366-75. [Crossref] [PubMed]
- Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 2012;107:1776-82. [Crossref] [PubMed]
- Popova T, Manie E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012;72:5454-62. [Crossref] [PubMed]
- Kraya AA, Maxwell KN, Wubbenhorst B, et al. Genomic Signatures Predict the Immunogenicity of BRCA-Deficient Breast Cancer. Clin Cancer Res 2019;25:4363-74. [Crossref] [PubMed]
- Connor AA, Denroche RE, Jang GH, et al. Association of Distinct Mutational Signatures With Correlates of Increased Immune Activity in Pancreatic Ductal Adenocarcinoma. JAMA Oncol 2017;3:774-83. [Crossref] [PubMed]
- Pitroda SP, Bao R, Andrade J, et al. Low Recombination Proficiency Score (RPS) Predicts Heightened Sensitivity to DNA-Damaging Chemotherapy in Breast Cancer. Clin Cancer Res 2017;23:4493-500. [Crossref] [PubMed]
- Zhao EY, Shen Y, Pleasance E, et al. Homologous Recombination Deficiency and Platinum-Based Therapy Outcomes in Advanced Breast Cancer. Clin Cancer Res 2017;23:7521-30. [Crossref] [PubMed]
- Shahda S, Timms KM, Ibrahim AA, et al. Homologous Recombination Deficiency in Patients With Pancreatic Ductal Adenocarcinoma and Response to Chemotherapy. JCO Precision Oncology 2018;1-11.
- Peng G, Chun-Jen Lin C, Mo W, et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat Commun 2014;5:3361. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017;23:517-25. [Crossref] [PubMed]
- Pishvaian MJ, Blais EM, Brody JR, et al. Outcomes in pancreatic adenocarcinoma (PDA) patients (pts) with genetic alterations in DNA damage repair (DDR) pathways: Results from the Know Your Tumor (KYT) program. J Clin Oncol 2019;37:191. [Crossref]
- Johnstone TC, Suntharalingam K, Lippard SJ. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem Rev 2016;116:3436-86. [Crossref] [PubMed]
- Agarwala SS, Kirkwood JM. Temozolomide, a novel alkylating agent with activity in the central nervous system, may improve the treatment of advanced metastatic melanoma. Oncologist 2000;5:144-51. [Crossref] [PubMed]
- Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355:1152-8. [Crossref] [PubMed]
- Golan T, Kindler HL, Park JO, et al. Geographic and ethnic heterogeneity in the BRCA1/2 pre-screening population for the randomized phase III POLO study of olaparib maintenance in metastatic pancreatic cancer (mPC). J Clin Oncol 2018;36:4115. [Crossref]
- Golan T, Kanji ZS, Epelbaum R, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer 2014;111:1132-8. [Crossref] [PubMed]
- Oun R, Moussa YE, Wheate NJ. The side effects of platinum-based chemotherapy drugs: a review for chemists. Dalton Trans 2018;47:6645-53. [Crossref] [PubMed]
- Jain A, Agostini LC, McCarthy GA, et al. Poly (ADP) ribose glycohydrolase can be effectively targeted in pancreatic cancer. Cancer Res 2019;79:4491-502. [Crossref] [PubMed]
- Parasido E, Avetian G, Naeem A, et al. The sustained induction of c-Myc drives nab-paclitaxel resistance in primary pancreatic ductal carcinoma cells. Mol Cancer Res 2019;17:1815-27. [Crossref] [PubMed]
- Tiriac H, Belleau P, Engle DD, et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov 2018;8:1112-29. [Crossref] [PubMed]
Cite this article as: Golan T, Brody JR. Targeting homologous recombination addicted tumors: challenges and opportunities. Ann Pancreat Cancer 2020;3:6.


