
Analysis of the receptor for advanced glycation end-products proteome and protein-protein interactome in PANC-1 cancer cells
Highlight box
Key findings
• Proteomic analysis of the effect of RAGE silencing in PANC-1 cells identified 166 proteins involved in three protein-protein interaction networks involved in ubiquitin and p53 pathways among others.
What is known and what is new?
• Ubiquitin and p53 are important regulators of cellular functions.
• RAGE-ligand binding can inhibit ubiquitination and promote p53, suggesting that RAGE may be a potential therapeutic target for pancreatic adenocarcinoma.
What is the implication, and what should change now?
• RAGE inhibitors may provide therapeutic benefit, and studies evaluating the effect of RAGE inhibitors in pancreatic cancer are needed.
Introduction
Each year an estimated 57,600 adults in the United States are diagnosed with pancreatic cancer, of which 90% are pancreatic adenocarcinoma (PA) (1). Around 80% of cases of pancreatic cancer are detected between the ages of 60–80 years with predominance in men and a higher rate of incidence and mortality in African, Japanese, and Anglo-American groups. PA can thus be categorized as a lethal disease facing clinicians with minimal non-surgical cures. The 5-year survival rates remain low at 10% highlighting the urgent need for therapies that enhance survival and improve outcomes (1).
The receptor for advanced glycation end-products (RAGE) is a member of the immunoglobulin superfamily as well as a pattern recognition receptor. RAGE has multiple functions, with the best characterized being to amplify and perpetuate the inflammatory response, other critical functions include protein transport, maintaining cell polarity, and promoting cell differentiation and cell division (2,3). The functions of RAGE appear to be dependent, in some ways, on the cell type or tissue bed expressing the receptor. Moreover, there are multiple isoforms of RAGE, including a full-length form needed for signaling, a N-terminal truncated form that appears to play a role in cell polarity in lung adenocarcinoma (4), and the soluble form of RAGE (sRAGE). Data from multiple studies show that RAGE is up regulated in diabetes, heart disease, and a variety of cancers (5-7).
RAGE likely plays a critical role in the pathogenesis of disease; however, its unique expression pattern suggests RAGE’s role is complicated. Studies have shown an increase in RAGE expression in adenocarcinomas in the pancreas and the lung (8,9). RAGE functions to promote tumor formation and the development of pancreatic lesions in murine models (10). Two ligands of RAGE-S100P and high mobility group box 1 (HMGB1) have high implications in promoting the role of RAGE in PA. S100P has been shown to both protect the PA cells from cytotoxicity and stimulate cell proliferation. In addition, S100P overexpression depict a fivefold increase in subcutaneous tumor growth while RAGE inhibition using siRNA has shown to inhibit tumor growth (11).
Apart from these, there are no studies depicting the impact of RAGE silencing on the proteome and the protein-protein interactions (PPI) in PA. The aim of the present study is to assess the role of RAGE in PA using gene silencing. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed following gene silencing and data subjected to PANTHER and STRING database analysis to classify proteins and their PPI networks, respectively. Three major PPI network clusters were identified. RAGE was found to contribute to p53 and ubiquitin pathways, which are both important for cell proliferation and maintenance. These data suggest that RAGE inhibitors could hold potential promise as therapeutics for PA.
Methods
Reagents
Unless otherwise noted, all reagents were purchased from Sigma-Millipore (Burlington, MA, USA).
Cell culture and gene silencing
Human PANC-1 cells (ATCC, Manassas, VA, USA) were grown in Dulbecco’s Modified Eagle Medium (DMEM) media containing 10% FBS and supplemented with penicillin/streptomycin. Cells were grown in a humidified 5% CO2 chamber at 37 °C temperature. Cells were grown in 6 well plates with an average of 2×105 cells/well in antibiotic free DMEM media supplemented with 10% fetal bovine serum (FBS). At ~60% confluence, all plates of cells were transfected with 6pmole siRNA of either RAGE or Scramble sequence siRNA (Santa Cruz, Dallas, TX, USA). After 48 hours, the cells were harvested. RAGE knockdown was then confirmed via PCR and western blot.
PCR
Total RNA was extracted using PureLink RNA mini-Kit (Thermo Fischer Scientific, Waltham, MA, USA) following the protocol of the manufacturer, and then reverse transcribed using the Superscript IV First-Strand synthesis system (Thermo Fischer Scientific). PCR was used to determine expression of RAGE mRNA. The primers used were ACT ACC GAG TCC GAG TCT ACC (RAGE forward), GTA GCT TCC CTC AGA CAC ACA (RAGE reverse). mRNA expression (DDCt) were then normalized to human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. Values presented are the mean of triplicate samples ± standard error (SE) performed from 3 independent studies.
Protein biochemistry
Following gene silencing, PANC-1 cells were rinsed with ice-cold PBS, lysed in RIPA buffer containing 1Å ~ protease and phosphatase inhibitors (Calbiochem, Burlington, MA, USA), and then cell lysates were subjected to electrophoresis on acrylamide gels. Following electrophoresis, the protein was transferred to Protran nitrocellulose membranes (Scheicher Schuell, St. Louis, MO, USA) for Western Blot analysis. Membranes were incubated with either goat polyclonal anti RAGE (Santa Cruz, sc-8229, 1:1,000), anti-p53 (1:1,000), or mouse polyclonal anti-ubiquitin (1:1,000) overnight. A secondary antibody (1:20,000) conjugate with horseradish peroxidase was applied, then incubated at room temperature for 1 h. The Supersignal West Dura (Thermo Fischer Scientific) was as used to develop a chemiluminescent signal that was captured using a UVP chemiluminescent imaging station (Thermo Fisher Scientific, Waltham, MA, USA) and compatible software. The data was normalized to β-actin (1:5,000) signal.
LC-MS/MS
LC-MS/MS was performed as previously reported (4,12). Briefly, proteins were solubilized in 50 mM Tris HCL/8M urea pH 8.0, vortexted, sonicated and then spun at room temperature for 10 minutes at 16,000 g. Proteins were then digested, reduced, and alkylated prior to LC-MS/MS. Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific) equipped with a nanoESI source was used for LC-MS/MS analysis. An Acclaim Pepmap™ 100 precolumn (Thermo Fisher Scientific) was used to elute digest peptide onto an Acclaim PepMap™ RSLC analytic column. Tandem mass spectra were searched against the human FASTA protein database from UniprotKB. The database was used to append for common contaminant proteins (e.g., human keratins obtained at ftp://ftp.thegpm.org/fasta/cRAP). All tandem mass spectrometry (MS/MS) spectra and proteins were identified with XCorr score cut-offs (13) and with 99% confidence through a reversed database search using the Thermo Proteome Discoverer 1.3 (Thermo Fisher Scientific). Peptide visualization and protein results were obtained using Scaffold v 4.3.4 (Proteome Software Inc., Portland, OR, USA), a program that relies on various search engine results (i.e., Sequest, X!Tandem, MASCOT) and which uses Bayesian statistics to reliably identify more spectra. The criteria used to identify proteins consisted of a minimum of two peptides identified at 0.1% peptide false discovery rate (FDR) and 90–99.9% protein confidence by the Protein Profit algorithm within Scaffold. Each experiment was repeated three times.
Proteome and Interactome database analyses
Accession numbers from differentially expressed proteins were uploaded into the PANTHER Classification System (Version 13.1 Released 2018-02-03) and STRING Interactome databases (Version 11). For PANTHER analysis, the ID list was uploaded, and accession numbers were searched against the human database to identify functional classifications. For protein -protein interactions, Accession numbers were uploaded to the STRING database, the full STRING network was used to show network edges with medium confidence. Clustering was performed using the Markov Cluster Algorithm (MCL algorithm) within the STRING database with an inflation factor of 3.
Statistical analysis
Student’s t-test (SigmaStat, San Jose, CA, USA) was performed for mean differences. Bonferroni’s correction was used for multiple testing in the PANTHER Overrepresentation Test and for post-hoc test for multiple comparisons. P<0.05 was considered significant.
Results
RAGE-dependent proteins in PANC-1 cells identified by LC-MS/MS
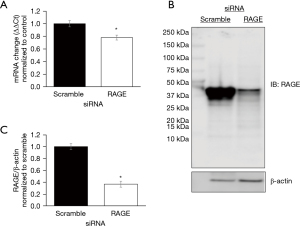
To assess the effect of RAGE in PANC-1 cells, we first confirmed reduction in RAGE mRNA and protein expression following gene silencing (Figure 1). Protein slurries were then subjected to proteomic analysis and 166 differentially expressed proteins were identified and then further classified using PANTHER classification. Proteins were classified according to their protein class, molecular function and biological function (Figure 2). A comparison of differentially classified proteins revealed that downregulated proteins consisted of 42 proteins involved in metabolic processes and 50 proteins involved in cellular processes. Additional functions among downregulated proteins included regulation, cellular organization, and response to stimulus functions (Figure 2A). Molecular functions included 37 proteins involved in binding and 26 proteins involved in catalytic activity (Figure 2B). Downregulated proteins also included proteins involved in nucleic acid binding, oxidoreductases, and lyase and transporter protein classes (Figure 2C).
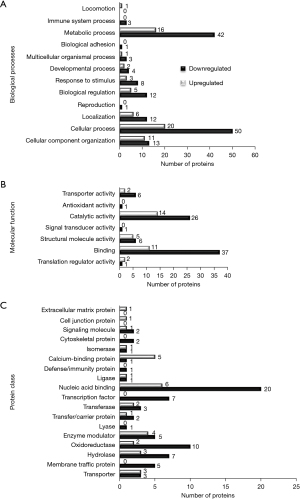
Gene expression predictions from the down-regulated proteins were completed. Table S1 details the overrepresentation tests from PANTHER showing fold enrichment changes in the expression of genes involved in biological processes, molecular function, and protein class. Reactome pathways are provided in Table S2 with overrepresentation pathways consistently largely of genes involved in respiratory electron transport pathways and non-sense mediated decay pathways. Additionally, a comparison of pathways among differentially expressed pathways is included in Table S3. These pathway comparisons demonstrated that the proteins involved in the ubiquitin proteosome pathway, p53 feedback loops 2 pathway, and p53 pathway were downregulated.
Western blot for p53 and ubiquitin
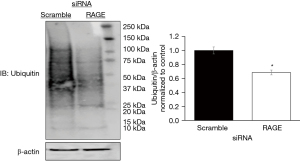
PANTHER and STRING databases identified the ubiquitin proteosome pathway, p53 feedback loops 2 pathway, and p53 pathways with RAGE silencing. We evaluated the effect of RAGE silencing on p53 and ubiquitin expression in PANC-1 cells. RAGE silencing reduced ubiquitin protein expression by 30% (Figure 3) and p53 protein expression by 70% (Figure 4).

STRING Interactome analysis derived PPI network with 3 clusters
STRING Interactome mapping analysis was performed for all differentially expressed proteins to identify their associated partners and PPI networks (Figure 5). Three protein clusters were identified from the computed predication of PPI networks, 1 large (II), and 2 small (I, III) protein clusters. In addition, the function(s) of identified proteins within the clusters are reported from the STRING database.
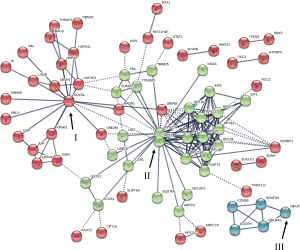
The large protein cluster (II) contains proteins RPL10, RPL21, RPL29, RPL31, RPL36, RPS18, RPS25, RPS27, RPL23A, and RPS11 which are actively involved in forming the 60S and 40S ribosomal subunits within the cell. In addition, protein interactions involving RPS27A, SUMO2, TRIM25, SQSTM1, and UBE2K are part of this cluster (II) and they play an active role in ubiquitination pathways within the cell. Other categories of proteins identified in this cluster involve ubiquitin-like proteins, and proteins involved in the autophagy, and proteins interactions pertaining to the TP53-binding protein-1.
The smaller protein cluster—cluster (I) involves proteins pertaining to transcriptional regulations and modifications. CDC5L, NUP153, LSM8, and SRSF5 are among the prominent proteins of the cluster affecting various pathways and proteins as shown in Figure 5. The other small protein cluster—cluster (III) involves proteins pertaining to the formation of proteins required in the Electron Transport Chain—mainly cytochrome c oxidase and NADH dehydrogenase.
Discussion
RAGE is abundantly expressed in the lung as well as a variety of epithelial malignancies (3,14); however, its expression and effects on tumorigenesis, especially in PA remains unclear. Recent work from our laboratory has identified that RAGE plays a crucial role in the regulation of several protein networks especially regulatory proteins like p53 and ubiquitin (4,12).
We and others have previously confirmed the presence of RAGE in PANC-1 cells, and that the over expression of RAGE in PA has been shown to have an incremental effect on cell proliferation (10). We silenced RAGE expression to study its effects on cellular processes. We identified three main functional protein clusters that were affected by RAGE with one cluster showing interactions with RAGE and ubiquitination proteins and processes. It has been previously reported that RAGE affects the ubiquitin proteosome pathways to down regulate proteins in murine glomerular mesangial cells (15), however its effect on the pathway itself in PA is not consistently reported. Proteomic analysis demonstrated a reduction in genes involved in the ubiquitin proteosome pathway with RAGE knockdown (Table S3). We observed a reduction in total protein ubiquitination following RAGE knockdown. Collectively, these findings suggest RAGE may affect several crucial cellular processes like nuclear transport, DNA replication and repair, and mitosis.
A key regulatory protein of mitosis is p53. Proteomic analysis demonstrated a reduction in p53 pathway proteins with RAGE knockdown (Table S3). This finding was confirmed by Western Blot Analysis which revealed that p53, a highly conserved protein in the pathway, expression was reduced by 70% reduction with RAGE knockdown (Figure 4). This observation suggests that RAGE contributes to several crucial cellular processes like DNA replication and repair, and mitosis. Indeed, this observation is supported by previous work demonstrating RAGE’s role in cellular differentiation and repair (3,6).
Data show that mutant p53 is associated with poorer prognosis (16) and chemoresistance (17) contributing to cancer aggressiveness in PA. However, the interaction between RAGE and p53 in adenocarcinoma is unclear. Studies show that RAGE ligand binding contributes to adenocarcinoma of the colon through JAK/STAT pathways, however, the role in PA is unclear (18). Mutant p53 has been extensively documented in PANC-1 cells (19-21) and RAGE-ligand binding may promote cell proliferation thought JAK/STAT pathway. In addition, RAGE has numerous ligands and specificity of the ligand may be important for the cancer progression.
Several additional process pathways such as the Ras pathway and oxidative stress response pathways were also downregulated with RAGE knockdown. Ras signaling pathways are crucial mediators of several malignant characteristics of transformed cells and are targeted for cancer therapy (22). Thus a downregulation in the Ras pathway would substantially affect the progression of tumorigenesis of PA. Nrf2 mediated oxidative stress response pathways are crucial to respond to oxidative stress and oxidative damage within the cell and thus are potential targets for cancer therapeutics. A downregulation of such oxidative stress response pathways through RAGE inhibition can thus also prove to be significant for cancer therapy. The JAK/STAT signaling pathway is activated in a large proportion of tumor solidification, cell migration, and proliferation (23). In PA, this pathway is shown to be a prominent marker and a potential therapeutic target for treatment (24). In this study, RAGE knockdown attenuated expression of JAK/STAT signaling pathway proteins (Table S3), suggesting that RAGE inhibitors could provide a potential benefit in mitigating JAK/STAT signaling in PA.
PA has been known to account for 90% of all pancreatic cancers and 5-year survival rates are low, primarily due to advance staging at diagnosis that results in a poor prognosis. RAGE has been known to play an important role in cell proliferation in PA (10). We used gene silencing coupled with proteomic databases and protein biochemistry to confirm a reduction in key regulatory proteins for cell proliferation like p53 and ubiquitin. These observations are aligned with others and suggest that RAGE plays an important role in a variety of cellular processes.
The current study has several limitations. First, only PANC-1 cells were used. Results from this study should be confirmed in other PA cell lines and biopsies of PA to validate findings as well as provide a more comprehensive understanding of the role of RAGE in PA. Second, we used gene silencing techniques and additional studies using more sophisticated approaches such as CRISPR allowing for gene editing could validate our findings. CRISPR would also provide an elegant method for identifying PPIs among critical proteins such as RAGE and p53.
Conclusions
PA remains a major source of cancer related morbidity and mortality with few treatment options that extend survival. As a result, there is a critical need for therapeutics for PA. The development of therapeutics requires advancing our understanding of oncogenesis of PA, as well as understanding the cellular and molecular complexities of PA. In this study we provide support for the role of RAGE in PA, specifically through p53 and ubiquitin pathways that participate in cell cycle progression and in protein expression. Our proteomic data suggest that RAGE may be a potential therapeutic target for PA, however, additional study is warranted.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at https://apc.amegroups.com/article/view/10.21037/apc-22-4/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://apc.amegroups.com/article/view/10.21037/apc-22-4/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Howard TJ. Pancreatic adenocarcinoma. Curr Probl Cancer 1996;20:281-328. [Crossref] [PubMed]
- Bierhaus A, Humpert PM, Morcos M, et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med (Berl) 2005;83:876-86. [Crossref] [PubMed]
- Brett J, Schmidt AM, Yan SD, et al. Survey of the distribution of a newly characterized receptor for advanced glycation end products in tissues. Am J Pathol 1993;143:1699-712. [PubMed]
- Downs CA. Analysis of RAGE Proteome and Interactome in Lung Adenocarcinoma Using PANTHER and STRING Databases. Biol Res Nurs 2021;23:698-707. [Crossref] [PubMed]
- Bi G, Yao G, Bian Y, et al. The Effect of Diabetes Mellitus on Prognosis of Patients with Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis. Ann Thorac Cardiovasc Surg 2020;26:1-12. [Crossref] [PubMed]
- Chen MC, Chen KC, Chang GC, et al. RAGE acts as an oncogenic role and promotes the metastasis of human lung cancer. Cell Death Dis 2020;11:265. [Crossref] [PubMed]
- Schraml P, Bendik I, Ludwig CU. Differential messenger RNA and protein expression of the receptor for advanced glycosylated end products in normal lung and non-small cell lung carcinoma. Cancer Res 1997;57:3669-71. [PubMed]
- Lv X, Qiao W, Leng Y, et al. Impact of diabetes mellitus on clinical outcomes of pancreatic cancer after surgical resection: A systematic review and meta-analysis. PLoS One 2017;12:e0171370. [Crossref] [PubMed]
- Liao YF, Yin S, Chen ZQ, et al. High glucose promotes tumor cell proliferation and migration in lung adenocarcinoma via the RAGE-NOXs pathway. Mol Med Rep 2018;17:8536-41. Erratum in: Mol Med Rep 2018;18:5302; Mol Med Rep 2021;24:720. [PubMed]
- Swami P, Thiyagarajan S, Vidger A, et al. RAGE Up-Regulation Differently Affects Cell Proliferation and Migration in Pancreatic Cancer Cells. Int J Mol Sci 2020;21:7723. [Crossref] [PubMed]
- Arumugam T, Simeone DM, Van Golen K, et al. S100P promotes pancreatic cancer growth, survival, and invasion. Clin Cancer Res 2005;11:5356-64. [Crossref] [PubMed]
- Downs CA, Johnson NM, Tsaprailis G, et al. RAGE-induced changes in the proteome of alveolar epithelial cells. J Proteomics 2018;177:11-20. [Crossref] [PubMed]
- Qian WJ, Liu T, Monroe ME, et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res 2005;4:53-62. [Crossref] [PubMed]
- Hofmann HS, Hansen G, Burdach S, et al. Discrimination of human lung neoplasm from normal lung by two target genes. Am J Respir Crit Care Med 2004;170:516-9. [Crossref] [PubMed]
- Huang KP, Chen C, Hao J, et al. AGEs-RAGE system down-regulates Sirt1 through the ubiquitin-proteasome pathway to promote FN and TGF-β1 expression in male rat glomerular mesangial cells. Endocrinology 2015;156:268-79. [Crossref] [PubMed]
- Nakamori S, Yashima K, Murakami Y, et al. Association of p53 gene mutations with short survival in pancreatic adenocarcinoma. Jpn J Cancer Res 1995;86:174-81. [Crossref] [PubMed]
- Fiorini C, Cordani M, Padroni C, et al. Mutant p53 stimulates chemoresistance of pancreatic adenocarcinoma cells to gemcitabine. Biochim Biophys Acta 2015;1853:89-100. [Crossref] [PubMed]
- El-Far AH, Sroga G, Jaouni SKA, et al. Role and Mechanisms of RAGE-Ligand Complexes and RAGE-Inhibitors in Cancer Progression. Int J Mol Sci 2020;21:3613. [Crossref] [PubMed]
- Hagi-Sharifia Taghavi M, Davoodi J, Mirshahi M. The effect of wild type P53 gene transfer on growth properties and tumorigenicity of PANC-1 tumor cell line. Iran Biomed J 2007;11:1-6. [PubMed]
- Ozaki T, Nakamura M, Ogata T, et al. Depletion of pro-oncogenic RUNX2 enhances gemcitabine (GEM) sensitivity of p53-mutated pancreatic cancer Panc-1 cells through the induction of pro-apoptotic TAp63. Oncotarget 2016;7:71937-50. [Crossref] [PubMed]
- Mohiuddin M, Chendil D, Dey S, et al. Influence of p53 status on radiation and 5-flourouracil synergy in pancreatic cancer cells. Anticancer Res 2002;22:825-30. [PubMed]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003;3:11-22. [Crossref] [PubMed]
- Thomas SJ, Snowden JA, Zeidler MP, et al. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer 2015;113:365-71. [Crossref] [PubMed]
- Pang C, Gu Y, Ding Y, et al. Several genes involved in the JAK-STAT pathway may act as prognostic markers in pancreatic cancer identified by microarray data analysis. Medicine (Baltimore) 2018;97:e13297. [Crossref] [PubMed]
Cite this article as: Doshi C, Johnson NM, Downs CA. Analysis of the receptor for advanced glycation end-products proteome and protein-protein interactome in PANC-1 cancer cells. Ann Pancreat Cancer 2023;6:5.


