Modeling human pancreatic ductal adenocarcinoma for translational research: current options, challenges, and prospective directions
Introduction
While overall cancer mortality has been decreasing for the past 30 years, the absolute mortality from pancreatic cancer continues to increase and the disease remains an outlier against the overall trend. Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic malignancy and one of the most lethal malignancies worldwide (1,2). By 2030, PDAC is expected to surpass colorectal cancer as the second leading cause of cancer-related deaths in the United States (3). The 5-year survival rate for PDAC remains a dismal 7–8% (4). At diagnosis, approximately 50% of patients have metastatic disease (5). PDAC most frequently metastasizes to the lungs, liver, and peritoneal cavity (6). With therapeutic intervention, the median survival for unresectable patients is between 6 and 11 months. Surgically resected patients have a longer median survival between 25 and 28 months (7). Although patients who are resected have a longer median survival, they represent less than a third of newly diagnosed patients (5). Most patients undergoing surgical resection ultimately have disease recurrence, with up to half of recurrences happening within the first year after resection (1). Lack of specific symptoms and a lack of an early screening test contribute to the advanced disease stage of most newly diagnosed patients (8,9).
Current systemic standard of care treatment options offer limited therapeutic efficacy (10). Patients are currently treated with standard chemotherapeutic regimens despite the heterogeneity seen in PDAC tumor response (1). Current standard regimens include gemcitabine/nab-paclitaxel and FOLFIRINOX (leucovorin, 5-fluorouracil, irinotecan, oxaliplatin) (11). A previous study in metastatic colorectal cancer suggested patients with certain mutations, including the ubiquitous mutation in PDAC, KRAS, do not benefit from certain drugs currently given in treatment plans (12). While KRAS is the most common driver mutation in PDAC, individual tumors behave differently, particularly in their response to chemotherapy (13). Recognizing this biological heterogeneity is important when evaluating an evolving approach to precision therapies.
Modeling PDAC in a research setting is challenging because it is difficult to represent the complex microenvironment ex vivo. Pancreatic cancer is uniquely paucicellular in the setting of a dense stroma with tumor cells having significant interactions with other components of the microenvironment including cancer-associated fibroblasts (CAFs), immune cells, neurons, microvasculature, and the extracellular matrix (14). Table 1 describes current challenges in accurately modeling PDAC (15,16). This manuscript describes the options in human models of PDAC and discusses how these models can be applied to translational research with an eye towards the clinical care of patients stricken with this disease.
Table 1
| Disease biology | Tissue acquisition | Heterogeneity | Tumor microenvironment |
|---|---|---|---|
| 1. KRAS mutation | Need tissue masses from a diverse population | 1. Intratumoral and intertumoral heterogeneity are challenges in representing PDAC | 1. Tumor microenvironment consists of fibroblasts, immune cells, and dense extracellular matrix |
| 2. Tumor suppressor deactivation and oncogene activation | 1. Endoscopic ultrasound fine needle biopsy | 2. Heterogeneity provides tumors with adaptability | |
| 3. DNA damage repair pathways | 2. Surgical specimen | ||
| 4. Angiogenesis |
The heterogeneity of PDAC makes it a challenging tumor to model since it is difficult to represent all tumor components ex vivo. PDAC is comprised of tumor ductal cells as well as a heterogeneous tumor microenvironment that should be considered when making decisions about modeling. (I) PDAC carcinogenesis is a multi-step process that is associated with multiple mutations and dysregulation of key cellular processes. Angiogenesis is also an important contributor to tumor progression. (II) Human tissue availability is not equal across institutions. Acquired tissue should represent a diverse patient population and include patients with early, middle, and late stages of disease. (III) There are intra-tumoral and inter-tumoral heterogeneity differences in tumors. Tumor cells can show different genotypic and phenotypic profiles. This heterogeneity impacts perceived efficacy when screening new therapies (15). (IV) Up to 90% of the tumor microenvironment can consist of dense extracellular matrix, immune cells, and fibroblasts. Modeling PDAC ex vivo accurately can help in understanding contributors to the tumor microenvironment and can impact clinical decision making. PDAC, pancreatic ductal adenocarcinoma.
Methods
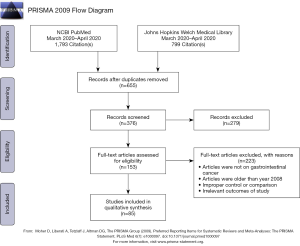
A comprehensive database review was undertaken of the NCBI PubMed, Google Scholar, and Johns Hopkins Welch Medical Library (powered by SCOPUS) resources for peer-reviewed manuscripts with search terms including: pancreatic cancer, PDAC, co-culturing, co-culturing PDAC, co-culturing fibroblasts, co-culturing lymphocytes, PDAC immunotherapy, KRAS mutations, KRAS PDAC, pharmacotyping, preclinical models, organoids, and xenografts. In total, 376 unique records were identified by our comprehensive review. Screening of these articles demonstrated the ubiquity of the use of human-derived models in PDAC cancer-biology research, particularly two-dimensional (2D) models of disease. Figure 1 shows the PRISMA flow chart accounting for this methodology (17). Initially, only articles related to pancreatic cancer were included, but the search was later expanded at the investigator’s discretion to other gastrointestinal malignancies. A secondary search was also done by review of manuscript references from the primary articles originally sourced.
Results
2D human cell lines
The first 2D cancer cell line was established at Johns Hopkins University in 1951 from a patient with cervical carcinoma (18,19). The techniques and ethical considerations from this experience have been discussed at length in both the research and lay press. The first pancreatic cell line was established almost 10 years later in 1963 (20). This technological advance changed the field of cancer biology research in fundamental ways. An immortalized cell line with stable phenotype and with the capacity to propagate on a culture plate gave rise to a plethora of novel reproducible research methods to study molecular biological processes as they relate to cancer (19). Since the 1960s, 2D cell lines have helped scientists understand how tumor cells arise, proliferate, migrate, and ultimately metastasize. Other advantages of using 2D PDAC cell lines are that they can be easily grown in large quantities, cultured quickly, transplanted into mice as xenografts, and, in the modern era, subjected to genetic screening (14).
2D cell lines have been studied extensively to understand tumor cell biology. Human pancreatic cancer (HPAC) was amongst the first lines to be widely used in contemporary research and was established by Gower and colleagues in 1985. Notably, the line was often used with a translational focus seeking data on unique therapeutic sensitivities in pancreatic cancer. It was the first PDAC line discovered to have a functional glucocorticoid receptor which, when treated with glucocorticoids, abrogated proliferation of the cell line. This finding suggested that glucocorticoids may have a functional role in the growth of pancreatic cancer tumors and set the stage for future cell-line based drug sensitivity work in PDAC (21). While this work failed to account for the inherent heterogeneity in PDAC disease biology or therapeutic response, it would evolve into translational work supporting the current chemotherapeutic backbones of systematic PDAC.
As molecular biology work shifted to a focus on genetic drivers of disease, 2D cell lines played a pivotal role in studying PDAC. This is largely because the dense stromal nature of primary human PDAC limited the yield of direct tumor sequencing in the early days of the Sanger method. Therefore, 2D cell lines served as the prototypical model of human PDAC and were used in initial efforts to identify the core signaling pathways in PDAC and lower-frequency mutations that represented potential therapeutic targets in this disease (22). It was from this work that the unique genetic signature in PDAC was appreciated, as cell lines were found to have almost universal KRAS mutations alongside a heterogeneous profile of associated mutations. The heterogeneity in associated mutations eventually lead to the hypothesis that, outside the discovery of a KRAS targeted therapy, heterogeneity could be expected in a tumor response to cytotoxic therapy (23). As a deeper appreciation was gained between the chemotherapeutic sensitivity in ex vivo models and the clinical response of a tumor, the study of 2D cell lines helped to usher in the era of multi-agent chemotherapy. Fountzila et al., for example, compared the effect of single agent exposure and drug combinations on the proliferation of MIA PaCa-2 cells and noted an additive inhibitory effect after cells were exposed to both Vincristine and Alizarin (24). Similarly, there was an additive inhibitory effect after cells were exposed to Vincristine and Adriamycin 6 hours apart (24). Ultimately, the concept of therapeutic synergy in multi-agent chemotherapeutic administration has resulted in the routine use of gemcitabine/nab-paclitaxel, or alternatively 5-fluorouracil/irinotecan/oxaliplatin, as standards of clinical care in pancreatic cancer.
Though 2D cell lines can be used for genomic and sequencing investigations, over the past several decades there remains a limited number of well-established and commonly used patient-derived PDAC lines in translational research. Table 2 shows seven cell lines that are currently available through the American Type Culture Collection (ATCC) (21,24-31). To diversify available cell lines, it is also possible to establish primary cell lines from patients presenting to academic medical centers, but procedural volume is varied across centers, patient consent must be obtained, and successful primary cell line establishment can be challenging (32).
Table 2
| Cell line | Year of origin | Organism | Person of origin | Derivation | Genes expressed | Genetic status of cell line | Significance | Differentiation |
|---|---|---|---|---|---|---|---|---|
| HPAC | 1985 | Homo sapiens | 64 years, female, Caucasian | Head of pancreas in primary tumor | +: keratin; –: vimetin and chromogranin A | DU-PAN-2, AUA1, HMFG1, KRAS | First reported PDAC line to express glucocorticoid receptor and forms heterogenous polar epithelial cells | Moderate |
| BxPC-3 | 1986 | Homo sapiens | 61 years, female | Primary tumor | Mucin, pancreas cancer specific antigen, carcioembryonic antigen | TP53, SMAD4/DPC4 | Inhibited by erlotinib: inhibition of EGFR might be a promising treatment | Moderate to poor |
| AsPC-1 | 1982 | Homo sapiens | 62 years, female, Caucasian | From metastatic site: ascites | Mucin, pancreas cancer specific antigen, carcioembryonic antigen | KRAS, TP53, SMAD4/DPC4 | Acquired resistance to cisplatin | Poor |
| MIA PaCa-2 | 1975 | Homo sapiens | 65 years, male, Caucasian | Primary tumor | Human colony stimulating factor, plasminogen activator | KRAS, TP53, CDKN2A/p16, SMAD4/DPC4 | Sensitive to asparaginase | Poor |
| Capan-1 | 1974 | Homo sapiens | 40 years, male, Caucasian | From metastatic site: liver | Mucin, blood type A, Rh+, HLA A2, B13, B17 | KRAS, TP53, CDKN2A/p16, SMAD4/DPC4 | Cells resistant to 5-fluorouracil which is identical to primary tumor | Good |
| Panc 10.05 | 1992 | Homo sapiens | Male, Caucasian | Head of primary tumor | Cytokeratins 7+18 | KRAS | KRAS oncogene mutation at Codon 12 | Poor |
| PANC-1 | 1973 | Homo sapiens | 56 years, male, Caucasian | Pancreas carcinoma of ductal cell origin | Glucose-6-phosphate dehydrogenase | KRAS, TP53, CDKN2A/p16 | First reported line from adenocarcinoma of the exocrine pancreasGrowth inhibited by L-asparginase | Poor |
Examples of some human pancreatic cell lines currently being used in research and their 2D genetic profile. These lines are readily available on atcc.org (21,24-31). We report the establishment and characterization of five new pancreatic cancer cell lines (PaCaDD-43, -60, -119, -135, -137). 2D, two-dimensional.
The challenges of successful line establishment are likely a result of the dramatic changes in growth conditions that accompany a transition from the environment in vivo to ex vivo. The establishment of 2D cell lines, grown over a base of plastic, induce the loss of tumor cell heterogeneity and cell polarity. The fundamental architecture that supports tumor cell growth is altered as compared to the primary tumor in vivo (14). Further, cells that are repeatedly passaged may not accurately represent the primary tumor (33). Genetic drift in cell lines causes a different effect on morphology and gene expression in the passaged cell line as compared to the primary tumor. Thus, using the same line continuously over several decades may not truly represent the primary tumor.
A large portion of PDAC primary tumor mass is comprised of a dense fibrotic stroma which is not represented in 2D culture. Culturing malignant ductal cells in 2D eliminates the ability to examine tumor-stromal interactions as immune cells, fibroblasts, and adipocytes are not included in the culture (31). 2D PDAC cell lines provide accessible models for studying tumorigenesis and progression as well as characterization of malignant cells, but they do not mimic the complexity of pancreatic cancer. Further, 2D cell lines may be limited in their potential to evaluate therapeutic efficacy because they lack a representative tumor environment. Prospective uses of 2D cell lines are commonly utilized in modern scenarios to understand disease biology through changes in gene expression, mRNA splicing, and the biochemistry of cells. They are also easily adapted for studies investigating metabolism, viability, and proliferation (32).
Xenografts
A xenograft is a tissue transplanted from one organism to another. The cells can be implanted orthotopically or ectopically (commonly subcutaneously) (34). Patient-derived tissues from primary tumor biopsy or resection, as well as human 2D cell lines, can be implanted into immunodeficient mice. A common immunodeficient strain used in xenograft studies is the NOD scid gamma (NSG) mouse (35). The two types of xenografts are commonly termed patient-derived tumor xenografts (PDX) and cell line-derived xenografts (CDX). Since CDXs are often generated from established cell lines, they are limited by some of the same barriers to generalization as their monolayer 2D cell lines of origin such as loss of tumor cell heterogeneity and a questionable capacity to re-establish polarity (33). There is also a limited correlation between CDXs and primary tumor histology (36). The PDX model offers a more comprehensive way to represent human PDAC since it implants a piece of three-dimensional (3D) tissue directly from a primary tumor (37). Garber et al. showed that PDX models better represent response to therapeutic intervention than CDX models and posited this may be due to a tumor microenvironment comprised of more ‘native’ tissues carried over in the PDX model (38). Overall, the PDX model has generated intense interest in translational researchers as a potential patient-specific avatar in precision medicine efforts to identify unique therapeutic sensitivities (39).
There are several limitations to consider with xenografts. First, a PDX model typically requires significant amounts of starting tissue and can be resource intensive (40-42). Even with an aggressive primary tumor, it is not uncommon for PDX generation to require up to 6 months of growth and a series of mice (43). This timeline is not realistic for precision medicine initiatives when the goal is to evaluate therapeutic efficacy in real time to aid in clinical decision making (44). Second, the success rates for de novo PDX models can be lower than that of cell line establishment, an area driven by unknown factors. Third, Delitto et al. have described infiltration with murine stroma in PDX models (45). With immunodeficient mice, an additional limitation is that the PDX model does not accurately recapitulate the interactions with the immune component of the tumor microenvironment (46). The use of mice as in vivo avatars of disease biology is an increasingly studied topic. Xenograft models do offer an opportunity to evaluate the efficacy of pharmacological interventions in a more complex biological system than that which can be mimicked in two-dimensions. One notable limitation, however, is in the recognition of the role that the immune system may play in chemotherapeutic and small molecule therapeutic efficacy. Additionally, the study of immuno-oncologic interventions are not possible in these humanized models (44).
Spheroids
An ongoing challenge in tumor modeling is accurately representing the 3D architecture of the primary tumor (34). Attempts to maintain organs ex vivo have been ongoing since 1938 when a cat pancreas was kept viable using a perfusion pump (47). Years later, this led to the discovery and utilization of artificial 3D matrices to enable the development of polarized cellular structures derived from human tumors. Spheroids, or 3D cultures in an artificial matrix, are principally derived from existing 2D cell lines that are embedded and allowed to propagate in layers or domes of collagen and/or Matrigel®. These cells are allowed to self-organize and demonstrate cell-cell, cell-microenvironment, and cell-matrix interactions that are more varied and nuanced than their 2D culture equivalents (48). The model was developed and popularized to address several challenges in the culture space as it relates to cell polarity and 3D architecture. Established cell lines can be readily propagated in artificial matrices and tend to coalesce together in a manner that seems to approximate that visualized in the human disease. Advantages of this approach are that tumor cell interactions and polarity are preserved and hypothesized to more accurately recapitulate the in vivo condition with the potential for more accurate therapeutic sensitivity testing in the 3D architecture setting (49-51).
Since spheroids originate from 2D cell lines, much like CDXs, a disadvantage is that their clinical/in vivo relevance may be limited by concerns over clonal expansion, limited success of establishing 2D cell lines from human PDAC and the time it takes to establish well-growing lines (33). One hypothesis would posit that the more complex a model is, the more representative it may be of the in vivo tumor from which it’s derived. While PDXs have the benefit of a more complex biological system, they are often expanded in a heterotopic position with a stroma that is a hybrid of mouse and human derived components as well as a lack of an immune infiltrate. Organoids and spheroids can be established, expanded, characterized and manipulated more rapidly when compared with PDXs, but are also limited by a mimic of the in vivo surrounding tumor microenvironment. Also, previous studies suggest the 2D cell line derived spheroids no longer retain the capacity to accurately mimic the polarity that would have been seen in the primary tissue. For example, Tsai et al. showed this using PANC-1 as a monolayer cell line to generate spheroids and compared it to an organoid grown from a primary tumor. Data suggest that the apicobasal polarity evident in the organoids derived primarily from patient tumors was absent from the spheroid cultures derived from previously established 2D lines (44).
An evolving use of spheroids in PDAC is their use in co-culturing models in which 3D models are derived to study the interaction between cancer cells and components of the surrounding stroma. In general, single cell line organoid/spheroid generation is the mainstay of biomass expansion. Secondary co-culture methods typically arise after establishment of single cell-line components to date. An exception is the use of organoid establishment methods for tissue characterization before the first passage. This technique invariably balances epithelial cell establishment and growth with some degree of apoptosis/anoikis of the other components of the tumor microenvironment that are not supported by factors within the culture conditions or media used (52).
3D human organoids
From spheroids, a natural progression for modeling human PDAC is with 3D organoids. The term ‘organoid’ was coined in part from the ability to be “organ-like” and mimic the tumor environment of the system from which the primary tissue is derived. The presumed difference between spheroids and organoids, used by most translational researchers, is that organoids are established primarily from enzymatically or mechanically dissociated tumor specimens whereas spheroids are established from existing monolayer (2D) cell lines (33). Organoid cultures, similar to spheroids, have two basic components: a 3D scaffold or matrix through which primary tumor derived tissue can propagate (typically Matrigel® or collagen) and a nutrient-rich growth-factor medium (1). Organoids may be generated from either a fine needle biopsy or resected tumor specimen. Labs with extensive experience cite success rates between 75–95% for organoid generation from either surgical resection or endoscopic biopsy (1). Previously, organoids have been used to model the mammary gland, colon, liver, small intestine, and stomach (40,53-56).
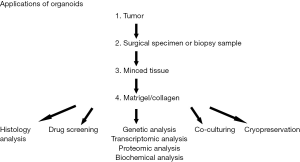
Similar to 2D cell lines, established organoids can be cryopreserved, stored long-term in liquid nitrogen, and regrown after thaw to reestablish culture (14). Patient-derived organoids (PDOs) are thus a model with a high enough success rate that it is a reasonable approach to consider to supplement fixed tissue biobanking efforts (1). An additional advantage to tumor organoids is that they can be passaged indefinitely for continuous study and experimentation to validate findings. In the 3D model, an added resource is possible in that growth factor supplementation allows for organoids to be derived from normal, non-cancerous tissues. These can be grown in parallel with tumor organoids as a control to compare morphological similarities and differences, but importantly, are not immortalized and the culture is eventually lost (usually within the first 5–10 passages) (57). Figure 2 shows the different applications of organoids (1,58).
The molecular methods used to study organoids are similar to those previously developed to investigate genetic, proteomic, biochemical, and transcriptional profiles in 2D lines (58). Weeber et al. completed sequencing of organoids 2–3 months after generation from metastatic sites of 14 patients with colorectal cancer. The organoid success rate from the metastatic biopsies was 71%. Sequencing data revealed that the organoids shared 90% of the somatic mutations with the metastatic site from which they were derived. An analysis comparing the DNA profiling of the primary tumor and the resulting organoids demonstrated a correlation of 0.89 (59). Driehuis et al. similarly demonstrated comparable morphological, histological and genetic features between 30 PDO lines and corresponding primary tumors (11). PDAC organoids, similarly, have been demonstrated to have a mutational profile consistent with that discovered when sequencing primary disease. Organoid phenotype also appeared to accurately reflect the morphological appearance of PDAC primary tumor seen on final histopathologic analysis (60).
The clinical response of PDAC patients to chemotherapy treatment has been inconsistent and unpredictable (1,60). Leveraging ex vivo organoid growth and chemotherapy testing to inform clinical care is one putative avenue of precision medicine in PDAC. Ex vivo organoid drug testing, or pharmacotyping, has been evaluated in both colorectal cancer and PDAC as a tractable method of personalized drug screenings and dosage profiling (1,60-62).
The median time required to complete organoid establishment and pharmacotyping can be less than 2 months in PDAC and is dependent, in part, on the quantity and quality of tumor mass available for organoid establishment (11). In patient’s undergoing surgery, for example, the process can be completed in its entirety during the patient’s post-surgical recovery period to assist in the selection of adjuvant therapy. This approach demonstrates a clinical relevance that has not been demonstrated to date by other methods of patient-derived ex vivo culture methods. Further, the heterogeneity of a primary tumor can be preserved in early PDO establishment as van de Wetering et al. demonstrated using a study of mutational profiles in gastrointestinal cancers (63,64). This current new approach may facilitate the shift from population-based medicine to personalized medicine as a potential method to maximize treatment efficacy in difficult-to-treat gastrointestinal tumor types. The utility of organoids in PDAC research is broad with researchers leveraging the technique to study many aspects of disease biology. This includes research centering on gene expression, transcriptional regulation, epigenetics, metabolism, chemotherapeutic sensitivity and therapeutic resistance mechanisms. It is these last two that also give rise to an interest in leveraging organoid technology for precision approaches to therapy in this disease (64).
There are several limitations which remain when considering organoid based translational research and clinical utilization. First, PDO establishment from patients who have received neoadjuvant therapy remains difficult, likely owing to less viable tissue at the time of resection (33). Additionally, Matrigel®, collagen, and the nutrient-rich growth medium required for organoid culture can be cost-prohibitive. Matrigel® and collagen are also subject to composition variation between lots which require frequent testing to maintain consistency in experimental design and implementation (65). The difference in composition of reagents also makes comparison of data across different laboratory settings challenging. Certain components of the growth media are derived by conditioning media overlaid atop a genetically engineered cell line, introducing variability in the process there as well. These factors, including Wnt3a and R-spondin1, are essential components of the organoid growth medium and variations in level may influence cellular metabolism and signaling, an important consideration when planning experiments (66).
In comparison to 2D models of disease, organoid models are able to accurately recapitulate the 3D architecture of the primary tumor. In the past, epithelial cells have been the focus of organoid work in PDAC. In embedded organoid models, the immune component, endothelial cells, and fibroblasts have typically not been propagated in culture (67,68). Improving representation of tumor heterogeneity requires development of co-culturing methods to include stromal components.
Co-culturing
Up to 90% of total tumor volume in PDAC can consist of supportive tissue and components of the tumor microenvironment (69). Supportive tissue can consist of CAFs, native components of the pancreatic connective tissue, the lymphatic vasculature, immune infiltrative cells, and deposited stroma. Cancer cells in vivo communicate with different components of the tumor environment to further support tumor proliferation and migration. These interactions between the microenvironment and tumor cells occur through physical contact and paracrine signaling (70). The extracellular matrix provides structural support for the tumor and is composed of collagen, enzymes, and proteins. Besides mechanical support, the extracellular matrix is also remodeled and selected components are leveraged to support physiological repair and tumor development (71).
CAFs are central to PDAC stroma and contribute to the regulation of cytokines, tumor growth factors, and immune infiltrate into the pancreatic cancer tumor microenvironment (72). Prior studies showing the complex interaction between tumor cells and the microenvironment suggested CAFs can play either a tumor supportive or tumor suppressive role (73,74). More recent data suggest CAFs can recruit regulatory T-cells into the tumor (75). Bachem et al. showed myofibroblast-like activated pancreatic stellate cells release mitogens for the proliferation of the cancer cells. Additionally, studies of tumor organoids suggest that CAFs may enhance the proliferative capacity for adenocarcinoma (76).
In considering an optimal multi-component model system, data from Tsai et al. suggest CAF heterogeneity may be induced by ductal co-culture with organoids from primary tumor as evidenced by differential expression of smooth muscle actin, a characteristic of myofibroblast-like activated pancreatic stellate cells (44). Co-culturing CAFs with organoids increased CAF expression of tumor promoting factors in a manner that was distinct from CAFs not co-cultured with organoids (70). Biffi et al. demonstrated using organoid-CAF co-cultures that TGF-beta and IL-1 were critical regulators of CAF heterogeneity when cultured in close physical proximity to a tumor organoid (77). These data suggest that models embracing a 3D approach to growth, which maintains cell polarity and cell-environment interaction through physical contact, are particularly important for translational research progress in the future. Understanding the mechanisms of tumor-CAFs crosstalk and the impact on tumor proliferation may provide new opportunities to identify prospective targets in the tumor stroma to manipulate with therapeutic intent.
While CAFs are key contributors to the tumor microenvironment, there are other cell types that also play important roles and could be adapted for use in co-culture with ductal organoids. For example, PDAC tissues demonstrate evidence of an acquired immune suppression caused by the upregulation of multiple pathways (78). Understanding these pathways may help distinguish why the results of past immunotherapy trials in PDAC have been discouraging (79). Current studies are examining interactions of organoids with other components of the tumor microenvironment by evaluating chemokines, cytokines, and microRNAs (80,81). Some of this work is focused on tumor angiogenesis including vascular endothelial growth factor and platelet-derived endothelial cell growth (82,83). There have been few widely adopted co-culture models representing the tumor lymphatic vasculature and differential nutrient delivery ex vivo, so further studies are warranted to continue to develop our understanding of complex interactions within the tumor microenvironment (84). Finally, a comprehensive multi-component model allowing for fibroblasts, immune infiltrates and components of the lymphatic vasculature environment will likely mature in future to provide insights into the role of these components in tumor progression and immunosuppression (67,85).
Discussion
A dense desmoplastic stroma and cellular heterogeneity contribute to the challenges of effectively modeling PDAC. These are important considerations when selecting models to improve our understanding of tumor biology and develop new therapeutics. Current approaches identifying reliable biomarkers and drug discovery in pancreatic cancer are hindered by the inability to replicate the tumor with adequate fidelity and an absence of the capacity to robustly model the tumor microenvironment ex vivo. Table 3 compares the different cancer modeling systems discussed in this review.
Table 3
| Model system | Cost | Time | Success rate | Surgical specimen or biopsy | Therapeutic response |
|---|---|---|---|---|---|
| 2D | $ | + | High | Surgical specimen | Low |
| Xenograft: CDX | $$ | ++++ | Medium | Surgical specimen | Medium |
| Xenograft: PDX | $$ | ++++ | Medium | Surgical specimen | Medium |
| Spheroids | $$ | ++ | Medium | Surgical specimen | Medium |
| Organoids | $$ | ++ | Medium | Both | High |
Comparison of two-dimensional (2D) cell lines, xenografts, spheroids, and three-dimensional (3D) organoids according to cost, time, success rate of line establishment, proliferation potential from surgical specimen or biopsy, and therapeutic response. PDAC, pancreatic ductal adenocarcinoma; CDX, cell line-derived xenografts; PDX, patient-derived xenografts.
2D cell lines have been used extensively to identify crucial PDAC mutations and to assess core hypotheses about the molecular mechanisms driving human disease. Xenograft cultures have been leveraged to assess characteristics of the human disease in a 3D in vivo setting with a focus on mechanisms impacting the tumor microenvironment. Although an ex vivo model, 3D organoid cultures established from surgical resections and tumor biopsies are genetically comparable to the phenotype of the primary tumor and are a flexible model for molecular and functional studies. In fact, single-cell DNA and RNA sequencing has shown similarities between the organoids and primary tumors (14). PDOs have shown feasibility as a modality for precision medicine and pharmacotyping, and may be used to inform personalized treatment plans (62,86).
Modeling the tumor microenvironment in a comprehensive way requires further study to realize a comprehensive ex vivo model of PDAC; therefore, it is key to consider CAF and immune cell populations, their relative frequency in the tumor microenvironment, and the relationships between cell density and tumorigenicity. Immune cell representation is especially important in the study of the emerging field of immunotherapy and its adaption for use in PDAC (44). Future PDAC modeling will be increasingly complex and comprehensive, as there is increasing interest in stromal biology and recognition of its important role in tumorigenesis and tumor progression. In recognizing the paucicellular component of PDAC tumor mass is comprised of epithelial cells, it is becoming increasingly important to account for the stromal biomass in translational research initiatives. Expanding co-culture techniques is becoming more feasible as genomic technology is increasingly capable of dissecting heterogeneous cellular populations to single-cell resolution. The advancement in single-cell and spatial genomic technologies also provides new opportunities to characterize tumor-stromal interactions. This will provide insight into the role each cell plays in the overall tumor biology.
Acknowledgments
Funding: JWZ is supported by 5T32-CA009071. RAB is supported by the National Institutes of Health/National Cancer Institute (K08CA248710) and a Stand Up To Cancer-Lustgarten Foundation Pancreatic Cancer Interception Translational Cancer Research Grant (grant number SU2C-AACR-DT26-17). Stand Up To Cancer (SU2C) is a division of the Entertainment Industry Foundation and funding is administered by the American Association for Cancer Research, the scientific partner of SU2C.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apc-20-29). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Aberle MR, Burkhart RA, Tiriac H, et al. Patient-derived organoid models help define personalized management of gastrointestinal cancer. Br J Surg 2018;105:e48-60. [Crossref] [PubMed]
- Hidalgo M, Cascinu S, Kleeff J, et al. Addressing the challenges of pancreatic cancer: Future directions for improving outcomes. Pancreatology 2015;15:8-18. [Crossref] [PubMed]
- Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res 2014;74:2913-21. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Coleman SJ, Watt J, Arumugam P, et al. Pancreatic cancer organotypics: High throughput, preclinical models for pharmacological agent evaluation. World J Gastroenterol 2014;20:8471-81. [Crossref] [PubMed]
- Rawat M, Kadian K, Gupta Y, et al. MicroRNA in pancreatic cancer: from biology to therapeutic potential. Genes 2019;10:752. [Crossref] [PubMed]
- Neoptolemos JP, Palmer DH, Ghaneh P, et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet 2017;389:1011-24. [Crossref] [PubMed]
- Sener SF, Fremgen A, Menck HR, et al. Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985-1995, using the National Cancer Database. J Am Coll Surg 1999;189:1-7. [Crossref] [PubMed]
- Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]
- Ying H, Dey P, Yao W, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2016;30:355-85. [Crossref] [PubMed]
- Driehuis E, Van Hoeck A, Moore K, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A 2019;116:26580-90. [Crossref] [PubMed]
- Fakih MG. Metastatic colorectal cancer: current state and future directions. J Clin Oncol 2015;33:1809-24. [Crossref] [PubMed]
- Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med 2018;8:a031435 [Crossref] [PubMed]
- Feigin ME, Tuveson DA. Challenges and opportunities in modeling pancreatic cancer. Cold Spring Harb Symp Quant Biol 2016;81:231-5. [Crossref] [PubMed]
- Ramón Y, Cajal S, Sesé M, Capdevila C, et al. Clinical implications of intratumor heterogeneity: challenges and opportunities. J Mol Med (Berl) 2020;98:161-77. [Crossref] [PubMed]
- Tesfaye AA, Kamgar M, Azmi A, et al. The evolution into personalized therapies in pancreatic ductal adenocarcinoma: challenges and opportunities. Expert Rev Anticancer Ther 2018;18:131-48. [Crossref] [PubMed]
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097 [Crossref] [PubMed]
- Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses: IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain hela) derived from an epidermoid carcinoma of the cervix. J Exp Med 1953;97:695-710. [Crossref] [PubMed]
- Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer 2002;2:315-9. [Crossref] [PubMed]
- Dobrynin YV. Establishment and characteristics of cell strains from some epithelial tumors of human origin. J Natl Cancer Inst 1963;31:1173-95. [PubMed]
- Gower WR Jr, Risch RM, Godellas CV, et al. HPAC, a new human glucocorticoid-sensitive pancreatic ductal adenocarcinoma cell line. In Vitro Cell Dev Biol Anim 1994;30A:151-61. [Crossref] [PubMed]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801-6. [Crossref] [PubMed]
- Cheung HW, Cowley GS, Weir BA, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A 2011;108:12372-7. [Crossref] [PubMed]
- Fountzilas G, Gratzner H, Lim LO, et al. Comparative effects of selected drug combinations on the growth of a human pancreatic carcinoma cell line (MIA PaCa-2). J Natl Cancer Inst 1986;76:37-43. [PubMed]
- Rückert F, Aust D, Böhme I, et al. Five primary human pancreatic adenocarcinoma cell lines established by the outgrowth method. J Surg Res 2012;172:29-39. [Crossref] [PubMed]
- Lieber M, Mazzetta J, Nelson‐Rees W, et al. Establishment of a continuous tumor‐cell line (PANC‐1) from a human carcinoma of the exocrine pancreas. Int J Cancer 1975;15:741-7. [Crossref] [PubMed]
- Lu YY, Jing DD, Xu M, et al. Anti-tumor activity of erlotinib in the BxPC-3 pancreatic cancer cell line. World J Gastroenterol 2008;14:5403-11. [Crossref] [PubMed]
- Tan MH, Nowak NJ, Loor R, et al. Characterization of a new primary human pancreatic tumor line. Cancer Invest 1986;4:15-23. [Crossref] [PubMed]
- Tan MH, Ming Chu T. Characterization of the tumorigenic and metastatic properties of a human pancreatic tumor cell line (ASPC-1) implanted orthotopically into nude mice. Tumour Biol 1985;6:89-98. [PubMed]
- Kyriazis AP, Kyriazis AA, Scarpelli DG, et al. Human pancreatic adenocarcinoma line Capan-1 in tissue culture and the nude mouse. Morphologic, biologic, and biochemical characteristics. Am J Pathol 1982;106:250-60. [PubMed]
- Deer EL, González-Hernández J, Coursen JD, et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas 2010;39:425-35. [Crossref] [PubMed]
- Kapałczyńska M, Kolenda T, Przybyła W, et al. 2D and 3D cell cultures--a comparison of different types of cancer cell cultures. Arch Med Sci 2018;14:910-9. [PubMed]
- Baker LA, Tiriac H, Clevers H, et al. Modeling pancreatic cancer with organoids. Trends Cancer 2016;2:176-90. [Crossref] [PubMed]
- Moreira L, Bakir B, Chatterji P, et al. Pancreas 3D organoids: current and future aspects as a research platform for personalized medicine in pancreatic cancer. Cell Mol Gastroenterol Hepatol 2017;5:289-98. [Crossref] [PubMed]
- Guo S, Gao S, Liu R, et al. Oncological and genetic factors impacting PDX model construction with NSG mice in pancreatic cancer. FASEB J 2019;33:873-84. [Crossref] [PubMed]
- Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009;324:1457-61. [Crossref] [PubMed]
- Sharpless NE, DePinho RA. The mighty mouse: Genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov 2006;5:741-54. [Crossref] [PubMed]
- Garber K. From human to mouse and back: “Tumorgraft” models surge in popularity. J Natl Cancer Inst 2009;101:6-8. [Crossref] [PubMed]
- Knudsen ES, Balaji U, Mannakee B, et al. Pancreatic cancer cell lines as patient-derived avatars: Genetic characterisation and functional utility. Gut 2018;67:508-20. [Crossref] [PubMed]
- Peterson JK, Houghton PJ. Integrating pharmacology and in vivo cancer models in preclinical and clinical drug development. Eur J Cancer 2004;40:837-44. [Crossref] [PubMed]
- Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res 2003;9:4227-39. [PubMed]
- Kondo J, Endo H, Okuyama H, et al. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci U S A 2011;108:6235-40. [Crossref] [PubMed]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:1039-49. [Crossref] [PubMed]
- Tsai S, McOlash L, Palen K, et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 2018;18:335. [Crossref] [PubMed]
- Delitto D, Pham K, Vlada AC, et al. Patient-derived xenograft models for pancreatic adenocarcinoma demonstrate retention of tumor morphology through incorporation of murine stromal elements. Am J Pathol 2015;185:1297-303. [Crossref] [PubMed]
- Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer 2007;7:645-58. [Crossref] [PubMed]
- Carrel A, Lindbergh CA. The culture of organs. Am J Med Sci 1938;196:732. [Crossref]
- Fanjul M, Hollande E. Morphogenesis of "duct-like" structures in three-dimensional cultures of human cancerous pancreatic duct cells (Capan-1). In Vitro Cell Dev Biol Anim 1993;29A:574-84. [Crossref] [PubMed]
- Wen Z, Liao Q, Hu Y, et al. A spheroid-based 3-D culture model for pancreatic cancer drug testing, using the acid phosphatase assay. Braz J Med Biol Res 2013;46:634-42. [Crossref] [PubMed]
- Yeon SE, No DY, Lee SH, et al. Application of concave microwells to pancreatic tumor spheroids enabling anticancer drug evaluation in a clinically relevant drug resistance model. PLoS One 2013;8:e73345 [Crossref] [PubMed]
- Longati P, Jia X, Eimer J, et al. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 2013;13:95. [Crossref] [PubMed]
- Norberg KJ, Liu X, Fernández Moro C, et al. A novel pancreatic tumour and stellate cell 3D co-culture spheroid model. BMC Cancer 2020;20:475. [Crossref] [PubMed]
- Katano T, Ootani A, Mizoshita T, et al. Establishment of a long-term three-dimensional primary culture of mouse glandular stomach epithelial cells within the stem cell niche. Biochem Biophys Res Commun 2013;432:558-63. [Crossref] [PubMed]
- Ootani A, Li X, Sangiorgi E, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med 2009;15:701-6. [Crossref] [PubMed]
- Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011;141:1762-72. [Crossref] [PubMed]
- Huch M, Gehart H, Van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299-312. [Crossref] [PubMed]
- Huang L, Holtzinger A, Jagan I, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 2015;21:1364-71. [Crossref] [PubMed]
- Boj SF, Hwang CI, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324-38. [Crossref] [PubMed]
- Weeber F, van de Wetering M, Hoogstraat M, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A 2015;112:13308-11. [Crossref] [PubMed]
- Tiriac H, Belleau P, Engle DD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov 2018;8:1112-29. [Crossref] [PubMed]
- Ooft SN, Weeber F, Dijkstra KK, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 2019;11:eaay2574 [Crossref] [PubMed]
- Seppälä TT, Zimmerman JW, Sereni E, et al. Patient-derived organoid pharmacotyping is a clinically tractable strategy for precision medicine in pancreatic cancer. Ann Surg 2020;272:427-35. [Crossref] [PubMed]
- Verissimo CS, Overmeer RM, Ponsioen B, et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. Elife 2016;5:e18489 [Crossref] [PubMed]
- van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161:933-45. [Crossref] [PubMed]
- Vukicevic S, Kleinman HK, Luyten FP, et al. Identification of multiple active growth factors in basement membrane matrigel suggests caution in interpretation of cellular activity related to extracellular matrix components. Exp Cell Res 1992;202:1-8. [Crossref] [PubMed]
- Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell 2012;149:1192-205. [Crossref] [PubMed]
- Jacobetz MA, Chan DS, Neesse A, et al. Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut 2013;62:112-20. [Crossref] [PubMed]
- Straussman R, Morikawa T, Shee K. Tumor microenvironment induces innate RAF-inhibitor resistance through HGF secretion. Nature 2012;487:500-4. [Crossref] [PubMed]
- Neesse A, Michl P, Frese KK, et al. Stromal biology and therapy in pancreatic cancer. Gut 2011;60:861-8. [Crossref] [PubMed]
- Fiorini E, Veghini L, Corbo V. Modeling cell communication in cancer with organoids: making the complex simple. Front Cell Dev Biol 2020;8:166. [Crossref] [PubMed]
- Daley WP, Peters SB, Larsen M. Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci 2008;121:255-64. [Crossref] [PubMed]
- Feig C, Gopinathan A, Neesse A, et al. The pancreas cancer microenvironment. Clin. Cancer Res 2012;18:4266-76. [Crossref] [PubMed]
- Ijichi H, Chytil A, Gorska AE, et al. Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J Clin Invest 2011;121:4106-17. [Crossref] [PubMed]
- Rhim AD, Oberstein PE, Thomas DH, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 2014;25:735-47. [Crossref] [PubMed]
- Lunardi S, Lim SY, Muschel RJ, et al. IP-10/CXCL10 attracts regulatory T cells: Implication for pancreatic cancer. Oncoimmunology 2015;4:e1027473 [Crossref] [PubMed]
- Bachem MG, Zhou S, Buck K, et al. Pancreatic stellate cells--role in pancreas cancer. Langenbecks Arch Surg 2008;393:891-900. [Crossref] [PubMed]
- Biffi G, Oni TE, Spielman B, et al. Il1-induced Jak/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov 2019;9:282-301. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Johnson BA, Yarchoan M, Lee V, et al. Strategies for increasing pancreatic tumor immunogenicity. Clin Cancer Res 2017;23:1656-69. [Crossref] [PubMed]
- Karagiannis GS, Pavlou MP, Diamandis EP. Cancer secretomics reveal pathophysiological pathways in cancer molecular oncology. Mol Oncol 2010;4:496-510. [Crossref] [PubMed]
- Zhang D, Li L, Jiang H, et al. Tumor–Stroma IL1b-IRAK4 feedforward circuitry drives tumor fibrosis, chemoresistance, and poor prognosis in pancreatic cancer. Cancer Res 2018;78:1700-12. [Crossref] [PubMed]
- Rosenberg H, Oppenheim J. The 2006 Dolph Adams Award and the State of the Journal of Leukocyte Biology. J Leukoc Biol 2007;81:369-71. [Crossref] [PubMed]
- Balkwill F, Mantovani A. Inflammation and cancer: Back to Virchow? Lancet 2001;357:539-45. [Crossref] [PubMed]
- Longo V, Brunetti O, Gnoni A, et al. Angiogenesis in pancreatic ductal adenocarcinoma: a controversial issue. Oncotarget 2016;7:58649-58. [Crossref] [PubMed]
- Hwang RF, Moore T, Arumugam T, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res 2008;68:918-26. [Crossref] [PubMed]
- Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science 2019;364:952-5. [Crossref] [PubMed]
Cite this article as: Suri R, Zimmerman JW, Burkhart RA. Modeling human pancreatic ductal adenocarcinoma for translational research: current options, challenges, and prospective directions. Ann Pancreat Cancer 2020;3:17.