
EBF2 contributes to pancreatic cancer progenitor cell differentiation and tumor suppression
Introduction
Pancreatic cancer is the third leading cause of cancer-related mortality in the United States; the 5-year patient survival is about 8% (1) due to the late-stage disease diagnosis, tangible tumor resistance to therapy (2) and aberrant post-treatment tumor relapse (3). Despite extensive research, targeted therapies for pancreatic ductal adenocarcinoma (PDCA) patients thus far benefited only marginally due to recurring mutations and intratumor heterogeneity (1,4). The complex PDAC pathobiology is established predominantly through KRAS mutations and the associated cellular signaling that contributes to cell proliferation and dedifferentiation (5,6). Studies have also identified other contender genes and genomic disruptions in human pancreatic exocrine tumors classified patients into different subclasses based on tumor histotypes and cancer-specific chromosomal rearrangements (7-9). Although such classifications reasonably correlated with overall patient survival, the underlying genomic instabilities have not corroborated into phenotypic heterogeneity prevalent in PDAC tumors. The pancreatic and duodenal homeobox-1 (PDX1)-expressing cells differentiate into functionally different lineages in normal pancreatic development. However, the differentiation trajectory of PDX1-expressing cells in tumors is mainly constrained by unidentified molecular mechanisms (5,10), contributing substantial clonal plasticity. Therefore, targeting cancer stem cells (CSCs)-like phenotypes for normalizing progenitor cell differentiation is emerging as an anti-cancer strategy.
Intratumoral heterogeneity instigated by partial activation of lineage-specific signals and concomitant terminal cell differentiation arrest corroborate functional vulnerabilities in progenitor cell subsets (11). This phenomenon may lead to vulnerabilities in steep hierarchical tissue organization indispensable for cellular senescence, potentially contributing to enhanced pluripotency turnover (12,13) and the evolution of more aggressive cell subtypes. Moreover, it remains elusive whether tumor-associated cell differentiation arrest potentially contributes to the development of PDAC and any progenitor cell subset acquires strong translational signals to override normal cell proliferation to advance tumor growth. Therefore, analysis of different tumor cell subsets may identify dynamic clonal population fated for long-term tumor propagation. Emerging evidence suggests that differentiation of neoplastic stem and progenitor cells could efficiently ablate tumor cell proliferation facilitate tumor suppression. For example, cancer cell differentiation eliminates treatment-resistant acute myeloid leukemia cells (14) and retinoids, implicated in the differentiation of colorectal cancer cells, repressing tumor growth and metastasis (15). Yet another study have shown that adipogenic differentiation inhibits cancer cell evolution leading to healthy progenitor cell development (16).
Studies have identified aberrations in Ebf1 and Ebf3 protein functions in PDAC due to cancer-specific somatic mutations (17). Moreover, EBF2 signaling has been implicated in the regulation of PI3K signaling (18), an effector of KRAS, deregulated in about 50% of PDAC patients suggesting that EBF2 could potentially interact with tumor signaling. We found that following EBF2-expression, the 3D-organoid forming PDAC epithelial cells differentiate in vivo into morphologically distinct pancreatic compartments. We further identified that the EBF2-signaling accurately recapitulates differentiation processes in vitro in KRAS mutant mesenchymal PDAC cells. Therefore, we provide evidence that differentiation-associated signaling inhibits PDAC growth representing a potential target to improve cancer therapy. We present the following article in accordance with the ARRIVE reporting checklist (available at https://apc.amegroups.com/article/view/10.21037/apc-21-17/rc).
Methods
The cell line, patient-tumor-derived primary cell culture, PDAC stem cells, and imaging
Patient-derived PDAC cells, TC2742-2, were generated from tumor samples obtained with informed consent under an approved clinical protocol of the National Cancer Institute (NCI), National Institutes of Health (NIH). Tumor tissues were washed in Hank’s balanced salt solution (HBSS), immediately transferred into the primary cell culture medium containing RPMI-1640:DMEM (1:1; Invitrogen) with 5% human serum, 10 µg/mL human insulin (Sigma), 2 mM L-glutamine, 100 units/mL penicillin and 100 µg/mL streptomycin (Thermo Fisher Scientific). Tumor tissues were minced with a razor blade and incubated with collagenase IV (0.24% w/v; Sigma) in HBSS overnight at 37 °C. Samples were treated 5 min with ammonium chloride solution for red blood cell lysis and filtered through 100 µM filters (Corning). Cells were collected by centrifugation and cultured under standard mammalian cell culture conditions at 37 °C. Adherent proliferating cell clones were manually picked, pooled, and characterized at the Cell Production Facility of the Surgery Branch, NCI and kindly provided by Dr. John Wunderlich. The PDAC cell line, MIA PaCa-2, purchased from the American Type Culture Collection (ATCC) were used. The patient-derived cells and cell line were cultured in pancreatic cell culture medium containing RPMI-1640:DMEM (2:1) with 5% fetal bovine serum (FBS; Sigma), 10 µg/mL human insulin (Sigma), 2 mM L-glutamine, 100 units/mL penicillin and 100 µg/mL streptomycin. Cells and assay cultures were maintained at 37 °C and 5% CO2. Cells were tested for mycoplasma and rodent pathogens, and none used tested positive for any of the contamination. To collect PDAC stem cells, sub-confluent MIA PaCa-2 and TC2742-2 cells were treated with pancreatic stem cell culture medium containing RPMI:DMEM (5:1) with 5% FBS, 25 µg/mL ascorbic acid, 10 µg/mL insulin, B27 (1X; Invitrogen), epidermal growth factor (1X hEGF; Lonza) and fibroblast growth factor 8 (FGF8, 50 ng/mL, R&D Systems) overnight, the floating CSCs were collected from the medium by centrifugation. Cells were resuspended in the pancreatic stem cell culture medium and grown in ultra-low adhesion culture plates for 3–5 days for CSC expansion. Proliferating CSC clones were collected by centrifugation at 1,000 × g for 5 min. and cells were dissociated into single cells with 1X Trypsin (Invitrogen) treatment for 5–10 min at 37 °C. The cells were grown in adherent plates for time-lapse imaging of CSC division. For BrdU pulse-chase experiments (19,20), CSCs were grown in the medium containing 1 µM BrdU (Sigma) for three passages. Cells were washed with PBS, fixed with 70% ethanol and treated with 2 N Hydrochloric acid (Sigma) for 1 h. Washed cells were incubated overnight at 4 °C with anti-BrdU-FITC antibody (BD Biosciences) and treated with 4',6-diamidino-2-phenylindole (DAPI, 1:1,000; Invitrogen) containing PBS for 10 min to identify newly synthesized DNA in the daughter cells in asymmetric cell division. Fluorescent images of BrdU-labeled template DNA and DAPI labeled DNA content were visualized using Olympus FluoView 1000 confocal microscope. CSC clones were dissociated and mixed with 7% DMSO (Sigma) and froze following the standard procedures to prepare stocks.
Organoid culture and imaging
Three-dimensional pancreatic organoids were grown by seeding CSCs on growth factor reduced (GFR) Matrigel (BD Biosciences) coated plates and cultured in modified pancreatic organoid culture medium (21) containing RPMI:DMEM (5:1) with 5% FBS, 25 µg/mL ascorbic acid, 10 µg/mL insulin, 1% B27 (Invitrogen), 25 ng/mL noggin, 5 mM nicotinamide (Sigma) and 50 ng/mL FGF8. Organoids were visualized and acquired using Olympus IX71 microscope and CellSens software (Olympus). For immunofluorescence, 5–10 d old organoids (TC2742-2) or spheroids (Mia PaCa-2) were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 1 h at 4 °C. Next, organoids or cell clusters were washed in PBS, permeabilized in 0.5% Triton X-100 (Sigma) and blocked with blocking buffer containing 5% bovine serum albumin (BSA) in PBS for 1 h at RT. Samples were incubated overnight at 4 °C with primary antibodies in blocking buffer. Primary antibodies (Cell Signaling Technology) used were rabbit anti-pancreatic and duodenal homeobox 1 (PDX1; 1:400), mouse anti-pan-cytokeratin (CK; 1:400), rabbit anti-zonula occludens-1 (ZO-1; 1:400), rabbit anti-SRY (Sex determining region Y)-box 9 (SOX 9; 1:400) and rabbit anti-VE-cadherin (1:400). Slides were washed with PBST (1 X PBS containing 0.05% Tween 20; Sigma), incubated with the corresponding anti-rabbit or anti-mouse Alexa-Fluor-488 or Alexa-Fluor-568 secondary antibodies (1:1,000; Molecular Probes) in PBS, washed with DAPI (1:1,000) containing PBS and mounted with Fluoromount-G (SouthernBiotech). Negative controls were treated with rabbit or mouse IgG or secondary antibodies alone. Immunofluorescence was detected using an IX71 fluorescence microscope and CellSens software.
EBF2-expression and progenitor cell differentiation
CSCs were seeded on adherent plates and cultured in the pancreatic stem cell culture medium. After 24 h, the medium was replaced with 5% FBS containing RPMI-1640. For stable transfection, Entry cDNA EBF2-ORFs-pCMV6-plasmid vectors or ORFs-pCMV6- control plasmids (OriGene Technologies) were added to the Lipofectamine 2000 (Invitrogen) containing Opti-MEM medium (Invitrogen), and treated to the cell culture. After 12–18 h, the medium was replaced with RPMI-1640 containing 5% FBS and 1 µg/mL insulin and cultured for 48 h. Then G418 (500 µg/mL) was added to the medium and maintained for 10 days (added fresh medium to the culture every second day) for selection. Immunoblotting verified the expression of EBF2 in cell clones. After the selection procedure, cells were maintained in the RPMI:DMEM (5:1) medium containing 5% FBS, 1 µg/mL insulin, 2 mM L-glutamine, 100 units/mL penicillin and 100 µg/mL streptomycin (complete medium). Most EBF2-transfected MIA PaCa-2 cell clones were differentiated into adipose or endothelial-like cells, evident from morphology. However, such morphological changes were not apparent in the EBF2-transfected TC2742-2 cells in culture. Three morphologically identical adipose or endothelial-like cell clones were pooled together to generate an endothelial or adipose clonal line (subclones) used in the experiments, whereas MIA PaCa-2 control, MIA PaCa-2 polyclonal, TC2742-2 control, and TC2742-2 EBF2-cells contain all antibiotic selected stably transfected cells.
Cell colony formation assay
Control or EBF2-transfected cells were dissociated and plated in 24 well plates (1,000 cells/well for MIA PaCa-2 and 2,000 cells/well for TC2742-2) and grown in a complete medium for 5 days. Cells were treated with methanol and stained with 0.5% crystal violet. Colonies were counted using an automated colony counter (Synbiosis), and the representative wells were photographed.
Cell proliferation assay
The cell proliferation assay was carried out using the WST-1 reagent (Roche Diagnostics) as instructed by the manufacturer. Briefly, 500 MIA PaCa-2 or 1000 TC2742-2 control and experimental cells were plated in 96 well plates and grown with a complete medium for 7 days. Cell proliferation was assayed by adding the WST-1 reagent to live cultures for 3 h at 37 °C. Absorbance was measured at a detection wavelength of 480 nm on a plate reader (BioRad) on the days indicated in the results.
Cell migration
Control and EBF2-transfected cells were plated at a density of 1.5×105 cells/well in a CIM plate and grown in the complete medium. Cell migration was recorded for a maximum 72 h using the xCELLigence RTCA DP device (Acea Biosciences) and plotted as cell index values. A baseline plot was generated for each group using cells grown in a serum-free medium.
Oil Red staining
MIA PaCa-2 control and adipose differentiated cells were maintained in the complete medium for 6–8 days. Cells were washed with PBS and fixed with 4% cold paraformaldehyde for 15 minutes. After three washes in PBS, neutral lipids were stained with Oil red O (Sigma). Images were captured using the Olympus IX71 microscope and the CellSens.
Subcutaneous tumor growth and serial transplantation studies
Animal studies were performed under an approved project protocol (No. 1215011) granted by the University of Maryland Institutional Animal Care and Use Committee, in compliance with the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) guidelines for the care and use of animals. Mice (Foxn1nu), 4–5 weeks old, were purchased from the University of Maryland Veterinary Resources and housed in a pathogen-free animal facility under standard conditions with access to food and water ad libitum. Experiments were performed in male or female mice based on the gender of the cells. No statistical methods were used to predetermine the sample size, and no blinding of the experimental groups was conducted. The number of animals in each group was estimated based on our previous experience in s.c. transplantation models. Cells for injections were counted using a cell counter (BioRad). Limiting dilution experiments were performed using control cells to assess the number of cells required to induce tumor growth in 80% or more mice upon cell injection. Subcutaneous tumors were established by injecting 2×106 Mia PaCa-2 control, endothelial, adipose, or polyclonal cells in 150 µL PBS on the flank of male mice using 24 G needles. Similar injections of 5×105 control or EBF2-transfected TC2742-2 epithelial cells were performed on female mice. The time needed for overt tumor palpation was noted. Tumors were measured twice a week using digital calipers, and tumor volume was calculated using the formula w2 × l/2, where l is the length and w is the width (22). Mice injected with differentiated adipose cells were monitored for 3-months for tumor palpation. Data from two independent experiments were used to determine tumor growth in adipose and endothelial subclones. MIA PaCa-2 xenotransplantation studies were terminated early due to the exceeding tumor size in one control animal permitted in the study protocol. Representative mice, as well as end-of-study tumors, were photographed using a Sony Cybershot camera. Tumor weight was gauged only in the MIA PaCa-2-tumors because TC2742-2-EBF2-tumors contained liquefied tumor cortex with loose cells covering outer solid tissue.
Next, we performed serial transplantation of TC2742-2 control (n=4) and EBF2-tumors (n=4) in naive mice to gauge whether EBF2 affects CSC functions in actively growing tumors. Equal sized primary tumor fragments were implanted s.c. in small skin incisions on the flank of the nude mice. Incisions were sealed with surgical clips or Vetbond (3M). In a separate study, small and large MIA PaCa-2-EBF2-polyclonal tumors were implanted. Mice were monitored for three months to gauge secondary tumor growth.
Adipose cell phenotypic plasticity induced by PI3K-p110α-specific drug
Unlike EBF2-expressing endothelial and epithelial cells, non-tumorigenic adipose-like cells maintain high PI3K-p110α-expression. Therefore, we treated adipose-like cells with a PI3K-p110α-specific inhibitor, alpelisib (Novartis), for seven days to gauge the role of PI3K-p110α on cell phenotype. Control cells treated with DMSO or 5 µM alpelisib-treated cells (2×106) were injected s.c. into the right flank of male Foxn1nu mice (n=5 per group). Mice were monitored for three months for tumor growth. End-of-study mice, excised tumors, and lymph nodes were photographed using the Sony Cybershot.
Immunofluorescence
Organoids and PDAC cells were washed in PBS and fixed in 4% neutral buffered formaldehyde for 10 min at RT, washed in PBS, and permeabilized with 0.2% v/v PBS/Triton X-100 for 5 min at RT. Samples were then washed with PBS and blocked with 3% BSA (w/v) in PBS at RT for 1 h before overnight incubation with primary antibodies (Cell Signaling Technology) to rabbit anti-E-cadherin (1:200), rabbit anti-vimentin (1:100), rabbit anti-VEGFR2 (1:800), rabbit anti-c-Myc (1:500), rabbit anti-β-catenin (1:100) and rabbit anti-caveolin-1 (1:400) at 4 °C. Samples were washed in PBST and treated with anti-rabbit Alexa-Fluor-488 or Alexa-Fluor-568 secondary antibodies (1:1,000; Molecular Probes) in PBS in the dark. Negative controls were treated with rabbit IgG or secondary antibody alone. Samples were washed with DAPI (0.5 µg/mL) containing PBS and mounted with Fluoromount-G (SouthernBiotech). Immunofluorescence was detected using Olympus IX71 fluorescence microscope and CellSens.
Immunoblotting
Equal numbers of control and EBF2-transfected cells were plated for immunoblotting experiments. Cells were washed in ice-cold PBS, trypsinized and collected as pellets after centrifugation at 3,000 ×g for 5 min at 4 °C. The cell pellets were incubated in 1 X RIPA cell lysis buffer (Millipore) with protease, and phosphatase inhibitor cocktails (Sigma), for 20 min at 4 °C. Cell lysates were then centrifuged at 20,000 ×g for 15 min at 4 °C. Protein concentration was measured in the lysate using the Pierce BCA Protein Assay Kit (ThermoFisher Scientific). Total protein (10–75 µg) was subjected to SDS-PAGE in pre-cast Bis-Tris 4–20% gradient gels (Invitrogen). Proteins were transferred onto a nitrocellulose membrane and blocked with 5% milk (Bio-Rad) in 1X Tris-buffered saline (TBS) for 1 h at RT. Membranes were then incubated overnight with primary antibodies at 4 °C. The following antibodies were used: EBF2 (1:200; R&D Systems), c-Myc (1:250; Novus Biologicals), Pparγ, adiponectin, Fabp4, C/EBPα, fatty acid synthase (Fasn), Sox2, Klf4, Oct4A, Nanog, E-cadherin, Cldn1, Snail1, Slug, Pten, PI3K-p110α, Akt, phospho-AktT308, Phospho-AktS473, mTOR, Raptor, Rictor, p70-S6K1, 4E-BP1, β-catenin, cyclin D1, (1:1,000; Cell Signaling Technology), phospho-mTORS2448 (1:200; Cell Signaling Technology), Gsk3β (1:250; BD Biosciences) and phospho-Gsk3βS9 (1:1,000; Cell Signaling Technology). β-actin (1:5,000; Sigma) was used as a protein loading control. Membranes were then washed in 1 X TBS containing 0.05% Triton-X100 (TBST) and probed with appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h at RT. The following secondary antibodies were used: anti-sheep (1:500; R&D Systems), anti-rabbit (1:1,000, Cell Signaling) and anti-mouse (1:10,000; ECL). Protein expressions were visualized with ECL Western Blotting Substrate (Thermo Fisher Scientific) followed by film (Thermo Fisher Scientific) exposure. SeeBlue (Invitrogen) protein ladder was used to determine the size of the protein band.
RNA purification and quantitative real-time PCR
Total RNA was isolated from cells using TRIzol (Life Technologies) extraction followed by purification with the RNeasy Kit (Qiagen) as per the manufacturers’ instructions. After NanoDrop RNA quantification and quality analysis, samples were reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). Unamplified cDNA samples were subjected to real-time quantitative PCR (qRT-PCR) with TaqMan Universal PCR master mix (Applied Biosystems). TaqMan probe and predesigned primers (Applied Biosystems) were used to detect the expression of the following genes: SIRT1 (Hs01009006_m1), PRDM16 (Hs00922674_m1), VEGFA (Hs00173626_m1), VEGFC (Hs00153458_m1), vWF (Hs00169795_m1), SOX2 (Hs01053049_s1) and NANOG (Hs02387400_g1). Gene expressions were normalized with β-actin expression quantified using the following primer and probe set: 5'-GCGAGAAGATGACCCAGA-3' (forward); 5'-CCAGTGGTACGGCCAGAG-3' (reverse). 5'-FAM-CCAGCCATGTACGTTGCTA-3' (probe). Samples were assayed in duplicate in biological replicates obtained at different time points. Cycling was performed with a TaqMan 7500 Real-time plus PCR system (Applied Biosystems).
Isoproterenol treatment and adipose cell growth on Matrigel plates
MIA PaCa-2 control and differentiated adipose-like cells were cultured in a complete cell culture medium, with or without 10 µM isoproterenol hydrochloride (Millipore) for 3–8 h. Uncoupling protein-1 (UCP1)-expression was assayed using qRT-PCR in quadruplicate replicates using the Taqman primer and probe for UCP1 (Hs00222453_m1 and Hs01084773_m1; Applied Biosystems) as described above.
Single-cell suspensions of endothelial and adipose cells (n=4/group) were grown on GFR-Matrigel with the complete cell culture medium for 12 days and images were captured on days indicated in the figure using Olympus IX71 microscope and CellSens.
Microarray
Gene expression studies were performed at the University of Maryland Marlene and Stewart Greenebaum Comprehensive Cancer Center (UMGCCC) Microarray Core Facility. Total RNA from control and experimental cells was isolated and processed for microarray hybridization on a GeneChip® Human Gene 2.0 ST Array (Affymetrix) using the GeneChip WT Plus labeling kit (Affymetrix). Samples were hybridized according to the GeneChip Whole Transcript (WT) Expression Arrays protocol (Affymetrix) with a hybridization time of 16 h. Arrays were scanned using a GeneChip Scanner 3000 7 G with an autoloader and Command Console Software (Affymetrix). Raw data (CEL files) were normalized (Gene Expression Omnibus accession number GSE78014) using Expression Console software (Affymetrix), and a robust multi-array (RMA) probe-set summarization algorithm was applied to all data. RMA intensity values were subjected to Principal Component Analyses using the MSCL Analyst’s Toolbox (JMP Statistical Discovery Software). Data were analyzed for outliers. One-way ANOVA was applied to the data. P values and False Discovery Rate (FDR) were generated. Differential gene expression in endothelial cells compared to the controls was identified with a 10% or less FDR and an absolute ≥2 fold change. Differential gene expression in adipose and EBF2-expressing epithelial cells from controls was identified at P<0.05 and absolute fold change ≥1.5 or more. Using absolute fold changes, top differentially expressed genes in each group were identified and listed.
Tumor histology
Tumor tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Tissue sections (5-µm) were stained with hematoxylin and eosin at the histology and pathology facility of the University of Maryland School of Medicine. Images were analyzed and acquired using Olympus IX71 fluorescence microscope and CellSens.
Statistical analyses
All statistical analyses, except the microarray data, were presented as mean and s.e.m. The P value P<0.05 was considered a statistically significant observation. Cell proliferation was analyzed by One-way analysis of variance (ANOVA) with Duncan’s multiple range test. Cell colony formation, qRT-PCR, vimentin expression in cells and tumor weight were analyzed by unpaired t-test comparing the means of two groups of values. No animals were excluded from the in vivo tumor growth analysis. The number of animals is specified in each figure or figure legend. Two-tailed Fisher’s exact test was used to determine significant differences in tumor growth in control and experimental groups. The predetermined criteria for secondary tumor growth were tumor induction or no tumor induction from implanted primary tumor fragments.
Results
EBF2 promotes the differentiation of cancer progenitor cells
Due to the inconsistencies in reproducing CSC-markers in solid tumors (23), consistent with other reports (6), we optimized manual isolation of CSCs from PDAC cell lines as reported previously (20,21). The E-cadherin-expressing epithelial CSCs and vimentin-expressing mesenchymal CSCs were isolated from TC2742-2 or the KRAS mutant MIA PaCa-2 cell line (24), respectively (Figure 1A). The cells divided asymmetrically in culture (Figure 1B; Figure S1A,S1B); the single-cell culture of pancreatic epithelial-CSCs initiated 3-dimensional (3D) organoid growth in 3-7 days with marked cyst-like growth comprising an outer layer of epithelial cells as reported previously (21,25). In contrast, seeding of 10-25 cells in culture plates promoted differentiation of layered epithelial cells and expansion of pancreatic duct-like outgrowths (Figure 1C). Lineage analysis revealed the expression of the neoplastic and epithelial marker, pan-cytokeratin (CK), pancreatic ductal, exocrine and endocrine progenitor cell marker, the pancreatic duodenal homeobox gene-1 (PDX1) and the tight junction protein, ZO-1 in the organoids (Figure 1D). By contrast, MIA PaCa-2 CSCs expanded clonally without organoid formation. The cells expressed CK, PDX1, and the ductal lineage marker, SRY-box (Sox)-9. The ZO-1 was uniformly expressed in the cell clones, but the endothelial adhesion marker VE-cadherin was expressed relatively in a small number of cell clones (Figure S1C).
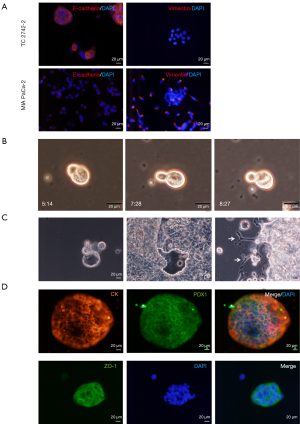
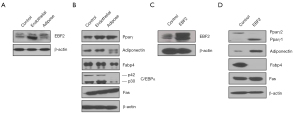
Previous studies have shown that PDAC growth originates from the ductal or acinar cells (26,27). Here we show that tumor-inducing PDAC stem cells spawn different pancreatic progenitors upon EBF2-expression, thus regulating cell fate decisions. The EBF2-expression in MIA PaCa-2 CSCs resulted in progenitor cell differentiation in vitro; most differentiated clones exhibited either endothelial or adipose-like morphology (Figure S2A). Because the single cell-derived clones differ in genetic and functional attributes (28), we pooled three clones of a specific lineage to generate a subline (hereafter referred to as endothelial and adipose subclones). Adipose clones were identified microscopically by large multilocular cells, a nuclear shift to one side of the cell and increased cytoplasmic lipid content (29). The NAD+-dependent deacetylase sirtuin-1 (SIRT1) decreased significantly in adipose-like cells suggesting decreased insulin sensitivity (30), whereas brown adipose-specific PR-domain containing protein-16 (PRDM16) marginally increased (Figure S2B,S2C). Notably, the PPARγ, a surrogate indicator of adipogenesis, upregulated, indicating reduced cellular quiescence, stemness and increased apoptosis (31). Conversely, control and adipose cells lacked detectable UCP-1-expression when treated with a pan-β-adrenergic agonist, isoproterenol. Consistently, adipose cell culture from EBF2-knockout mice did not induce UCP1-expression following adrenergic stimulation (32). Adiponectin, Fabp4, and the pluripotency enhancer CCAAT/enhancer-binding protein alpha (C/EBPα) (33) decreased with adipose-like cell differentiation (Figure 2A-2D).
Endothelial-like cells promoted vascular tube growth when grown on Matrigel plates. Under normal culture conditions, the cells displayed increased endothelial factors Vegfr-2, VEGFC and vWF (Figure S2D,S2E). Compared to the control and adipose group, endothelial differentiation manifested increased cell colony formation and migration, whereas cell proliferation was repressed responsive to differentiation (Figure S3A-S3D). In epithelial PDAC cells, EBF2-expression suppressed cell colony formation, proliferation and migration (Figure S3E-S3H). If cell proliferation is an indicator of tumor growth, our results predict decreased cell proliferation in Pparγ-expressing adipose, endothelial and epithelial cells could inhibit tumor growth.
Cell differentiation inhibits PDAC growth
We assessed the relative contribution of differentiated subclones on malignant progression by gauging time and rate of tumor palpation and growth. Compared to a previous report (34), we excluded Matrigel from in vivo tumor growth studies because others have reported that Matrigel induces endothelial cell proliferation from adipose tissue fragments (29). The single-cell culture of endothelial cells on Matrigel plates promoted clonal expansion, whereas adipose cells exhibited quiescence (Figure S4). In our limiting dilution studies, subcutaneous (s.c.) injections of 2×106 number of MIA PaCa-2 cells in male Foxn1nu (nude) mice substantially increased the percentage of mice with tumor growth compared to consecutively diluted cells. The control mice displayed a robust tumor growth (9/9); tumor palpation occurred on day 13 of tumor cell transplantation. By contrast, adipose cells abolished tumor formation (0/13) upon s.c injection in mice. Endothelial cells generated tumors in 73% (8/11) mice with a consistent delay (28 days) in tumor palpation; the tumor growth rate was significantly reduced. Next, to gauge the overall effect of EBF2 on tumor suppression, polyclonal cells containing all EBF2-transfected cell subclones were implanted into mice. In this group, 75% (6/8) of mice developed tumors; some tumors grew faster nexus endothelial growth characteristics while others displayed slow growth (Figure 3A-3C; Figure S5A). Serial transplantation of small polyclonal tumors failed to generate secondary tumors suggesting impaired CSC repopulation, whereas large polyclonal tumor fragments developed secondary tumors (Figure S5B).
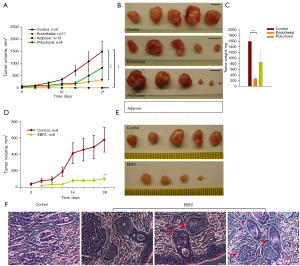
Next, injections of 5×105 PDAC epithelial CSCs (TC2742-2, control) in female Foxn1nu mice initiated tumor growth (8/8) on day 10, whereas similar injections of EBF2-expressing cells generated tumors only in 75% (6/8) of mice on day 20. Overall, tumor growth was repressed with EBF2-expression in cells, further confirming the anti-tumor effects of EBF2 on PDAC (Figure 3D,3E; Figure S5C). Notably, EBF2-tumors contained little solid tissue covering the inner interstitial fluid-filled cortical area. Histology of tumors revealed structural differentiation of epithelial cells into pancreatic ductal and acinar-like cells suggesting that EBF2 is essential for pancreatic tissue differentiation. Tumor histology indicated ductal-acinar structural distortion upon tumor expansion resembling early pancreatic intraepithelial neoplasia suggesting that other pivotal factors override tumor inhibitory signals of EBF2 (Figure 3F). Furthermore, the upregulation of retinol-binding protein-4 (RBP4) in EBF2-expressing epithelial cells suggests that retinol signaling potentially supports EBF2-induced differentiation processes. Conversely, primary tumor engrafts generated secondary tumors upon transplantation, indicating CSC exuberance in actively growing tumors (Figure S5D).
EBF2 regulates stemness and EMT signaling
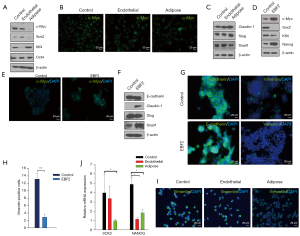
Next, we examined the molecular mechanism of tumor suppression in different PDAC subclones. In KRAS-mutant PDAC, MYC controls self-renewal and metastasis (2). Here, adipose-like differentiation clobbered c-Myc and the interacting snail-1-expression, whereas Klf4, a negative regulator of cell proliferation, activated (35) relating tumor suppression. Conversely, Klf4 downregulated and snail-1 upregulated in the endothelial and epithelial subsets. Notably, E-cadherin was consistent in both control and EBF2-expressing epithelial cells, even in the presence of Snail-1 activation (36). Furthermore, a meaningful decrease in vimentin-positive cells in epithelial clusters suggests that EBF2 maintains epithelial cell integrity by decreasing EMT processes. Claudin-1 (Cldn1) is consistently upregulated with EBF2-expression, and reduced levels of Cldn1 in epithelial cancers are associated with poor patient survival (37). In the adipose and EBF2-expressing epithelial cells, Sox2 repressed, suggesting a pivotal role of cellular differentiation in limiting tumor cell plasticity (38,39) and aggressive tumor behavior (40) (Figure 4A-4J). These results indicate tighter pluripotency and EMT signaling regulation by EBF2 in epithelial and mesenchymal-type PDAC cells.
EBF2 modulates cancer signaling through differentiation processes
To analyze cancer signaling upon EBF2 expression in cells, we studied PI3K-mTOR axis and β-catenin signaling (Figure 5). First, we investigated if the differentiation program influences KRAS effectors, and the relevance of PI3K signaling in the subclones. PI3K-p110α was highly expressed in control and adipose-like cells in contrast to a significant downregulation in EBF2-expressing endothelial and epithelial cells. Consistently, Akt is repressed in the endothelial and epithelial subsets, whereas decreased Akt-phosphorylation found in the adipose-like cells potentially contributes to tumor suppression.
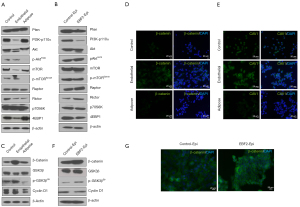
Furthermore, mTOR phosphorylation in adipose-like cells is strongly associated with low Akt-phosphorylation status. Rictor, a PI3K-dependent cell survival factor of mTORC2 signaling complex downregulated in the adipose and endothelial subclones, whereas the adipogenic activator, Raptor (41), and the mTORC1 downstream target p70S6K were unchanged. Concordantly, 4EBP1 upregulated in the adipose-like cells supporting decreased cell proliferation (42,43) and reversing metabolic dysfunction (44). In epithelial cells, activation of Rictor and Raptor could induce cell survival and tumor growth in the presence of activated c-Myc (45). In contrast to the adipose cells, EBF2 suppressed 4EBP1 in the epithelial and endothelial cells exhibiting a tissue-specific expression pattern in the tumor-producing subsets (Figure 5A,5B).
Rossi and Gonzalez reported that activation of Drosophila-p110 (dp110) in neural stem cells suppresses tumor growth (46). Although our observation is in contrast to other reports (47), it is noteworthy that most PI3K inhibitors repeatedly failed in clinical settings after an initial anti-tumor response (48). Treatment of non-tumorigenic adipose-like cells with a PI3K-p110α specific inhibitor, Alpelisib, induced transdifferentiation of cells into long spindle fibroblast-like phenotype but s.c. transplantation of cells did not generate tumors in most recipient mice. Whereas, a mouse in the treatment group showed axillary lymph node enlargement a week after tumor cell injection grew tumor at the site of cell injection. The PI3K-p110α and phospho-AktT308 increased in the Alpelisib treated adipose-like cells further substantiates Akt-phosphorylation and associated phenotypic plasticity, presumably reflecting the reactivation of oncogenic signaling (Figure S6A-S6E).
The β-catenin signaling activated with EBF2-expression in the epithelial subsets
To further elucidate the molecular mechanism associated with epithelial cell differentiation and tumor growth, we analyzed β-catenin signaling implicated in the process of pancreatic tissue development (49), vascular stability (50) and tumor growth (51). In the epithelial and endothelial cells, β-catenin increased with EBF2, presumably due to active GSK3β-phosphorylation, indicating that EBF2 preferentially affects epithelial functions. Conversely, in the adipose subset, GSK3β upregulated with a corresponding decrease in GSK3β-phosphorylation and β-catenin expression. The adipocyte membrane factor caveolin-1 (CAV1) (52) upregulated in the endothelial-like cells concomitant with β-catenin expression, presumably due to the interaction of the CAV1 scaffolding domain with β-catenin for transcriptional regulation (53) (Figure 5C-5G). Perhaps, β-catenin also influences downstream c-Myc and the differentiation suppressor, cyclin D1, in endothelial and epithelial cells upon EBF2-expression as these factors are repressed with adipose-like cell differentiation.
Therefore, our studies indicate that cell differentiation promotes substantial cellular and molecular heterogeneity that downplays key tumor proteins in PDAC. Additionally, at the transcription level, the tumor-promoting genes, ITGA3, PRND, and TSPAN1 repressed in the endothelial cells, whereas cell adhesion-related factors such as CDH10, CDH12, and the anti-apoptotic regulator, BIRC2, preferentially expressed. The adenocarcinoma-related factors KLK6 and TSPAN1, the matrix-related gene associated with poor PDAC patient survival COL1A2 (54), the adenosquamous pancreatic tumor-associated gene TP63 (55), COL5A2 and ITGB4 were repressed in the differentiated adipose cells. The regulator of fat metabolism NTS is substantially expressed in adipose-like cells. In the pancreatic epithelial cells, EBF2 activated tumor suppressor ADAM23, WNT suppressor CCDC88C, retinol carrier RBP4, and BMP antagonist SOSTDC1. In contrast, pancreatic cancer-associated genes DUOX2 and EDIL3 were repressed, substantiating the anti-tumor effects upon cell differentiation (Figure S7A-S7D).
Discussion
Transcriptome profiling of patient tumors and combination therapy studies suggests that intratumoral genetic and metabolic heterogeneity determines clinically relevant therapeutic endpoints in PDAC (56,57). We identify that EBF2 is indispensable for PDAC progenitor cell differentiation and tumor suppression. Epithelial-CSCs grew as 3D-organoids in modified culture conditions, expressing different pancreatic developmental markers necessary for lineage differentiation. Our data suggest that EBF2 promotes epithelial cell functions by activating E-cadherin, Cldn1, and β-catenin and inhibiting vimentin expression. Evidence suggests that in pancreatic cancer, E-cadherin activation represses tumor cell metastasis (58). EBF2 DNA binding motifs enriched within Pparγ binding sites promote Pparγ-expression and brown adipose differentiation (59). We found that the adipose lineage marker Pparγ activated in all PDAC subsets upon EBF2-expression as in normal adipose differentiation. Nonetheless, only a subset of mesenchymal cells differentiated as adipose-like cells with reduced colony formation and cell migration. A caveat in our studies is that although the EBF2-expressing endothelial (specialized type of epithelial cells) and epithelial cells express high β-catenin, unknown molecular mechanisms differentially regulated cell colony formation and migration in the cell types. Our results highlight that pancreatic tumor growth is likely caused by the proliferation of stem/progenitor cells with redundant differentiation signaling. Detailed studies are required to understand how different lineage-specific cues are activated under identical niche conditions for cell differentiation.
The EBF2-expression modestly repressed in many human adult tissues upon terminal differentiation (60). Retinoic acid is a critical regulator of pancreatic cell differentiation (61). In PDAC epithelial cells, the retinoid-binding protein RBP4 substantially upregulated with EBF2, drives pleiotropic differentiation of pancreatic tissue compartments upon s.c. transplantation. Nevertheless, our study does not corroborate the cellular and molecular processes involved in the stepwise progression of PDAC tumors, specifically acinar and ductal dissociation, ductal convolution, and cell degeneration, as evident from tumor histology. Presumably, lineage-associated signaling activated in the differentiation process plays a central role in PDAC tumor suppression. For example, activation of Cldn1, β-catenin and c-Myc, and downregulation of vimentin in EBF2-expressing epithelial and endothelial cells are epithelial-specific molecular traits. The vulnerabilities in β-catenin and c-Myc-expression mediate tumor formation (53,62,63), subdued levels of c-Myc, β-catenin, and phospho-GSK3β in adipose-like cells likely contribute to tumor abolition. Nonetheless, high expression of β-catenin in pancreatic exocrine cells possibly activates pancreatic progenitor cell proliferation and differentiation (64). Our results also suggest that low lipolysis and high cellular fat content in adipose-like cells are likely due to low cellular Fabp4-expression. The lack of expression of brown fat activator, UCP-1, in Pparγ-expressing cells with adrenergic stimulation suggests that cells are refractory to brown adipose differentiation. Overall, molecular mechanisms pertained to progenitor cell differentiation suppress malignant signaling.
In pancreatic cancer, PI3K signaling activates normal adipose cell differentiation (65) and drives tumor signaling in response to KRAS activation (66). In our study, expression of PI3K-p110α varied across the cell subsets analyzed, repressed in the endothelial and differentiating epithelial cells but, Akt-phosphorylation associated with tumor signaling activated supporting tumor growth. Stable expression of PI3K-p110α and decreased Akt-phosphorylation in adipose-like cells likely contribute to tumor suppression. Studies illustrate that inhibition of PI3K promotes tumor metastasis and growth (62), whereas signaling activation results in tumor suppression (46). Additionally, downregulation of core mTOR signaling and cyclin D1 in adipose cells support anti-tumor activity. Using different progenitor cell subsets with varying levels of PI3K-p110α expression, we confirmed that EBF2 is an upstream regulator of PI3K signaling; lack of EBF2-expression in adult CSCs adversely affects lineage decision, activating tumor growth. Additionally, Alpelisib-induced activation of Akt-phosphorylation resulting transdifferentiation of cells suggests that the PI3K-Akt signaling is attributed to differentiated adipose cell state.
In conclusion, we define that EBF2 mediates the differentiation of various pancreatic tissue types. A notable finding is that the epithelial cells attained lineage commitment in vivo in tumors upon EBF2 activation, but the differentiation signals are not enough to abolish tumor growth. Indeed, differentiation of adipose-like phenotype remodeled tumor signaling. Thus, further studies are warranted to identify the molecular mechanisms that eliminate tumor growth. A better understanding of the regulation of cell differentiation of pancreatic cancer progenitor cells could lead to the discovery of therapeutic options with broad implications.
Acknowledgments
We thank Dr. Richard Alexander for providing the drug BYL719 and Dr. Patrick Seale, University of Pennsylvania, for the mouse EBF2 plasmid for initial experiments not described in this paper. We thank Dr. Stuart Martin, UMGCCC, for confocal microscopy imaging facility access, Sayeh Ebrahimian for technical assistance, Jing Yin, UMGCCC, for microarray experiments, Dr. Susan G. Dorsey and Lassiter Cameron, University of Maryland School of Nursing and Mohammad Afnan Khan for technical support. We also thank Dr. William H. Matsui, Johns Hopkins University, for discussions and suggestions.
Funding: This study was supported by the Department of Surgery, University of Maryland School of Medicine.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://apc.amegroups.com/article/view/10.21037/apc-21-17/rc
Data Sharing Statement: Available at https://apc.amegroups.com/article/view/10.21037/apc-21-17/dss
Peer Review File: Available at https://apc.amegroups.com/article/view/10.21037/apc-21-17/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://apc.amegroups.com/article/view/10.21037/apc-21-17/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal studies were performed under an approved project protocol (No. 1215011) granted by the University of Maryland Institutional Animal Care and Use Committee, in compliance with the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Alistar A, Morris BB, Desnoyer R, et al. Safety and tolerability of the first-in-class agent CPI-613 in combination with modified FOLFIRINOX in patients with metastatic pancreatic cancer: a single-centre, open-label, dose-escalation, phase 1 trial. Lancet Oncol 2017;18:770-8. [Crossref] [PubMed]
- Ischenko I, Petrenko O, Hayman MJ. Analysis of the tumor-initiating and metastatic capacity of PDX1-positive cells from the adult pancreas. Proc Natl Acad Sci U S A 2014;111:3466-71. [Crossref] [PubMed]
- Viale A, Pettazzoni P, Lyssiotis CA, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014;514:628-32. [Crossref] [PubMed]
- Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013;369:1691-703. [Crossref] [PubMed]
- Puri S, Folias AE, Hebrok M. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell 2015;16:18-31. [Crossref] [PubMed]
- Grabocka E, Pylayeva-Gupta Y, Jones MJ, et al. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell 2014;25:243-56. [Crossref] [PubMed]
- Noll EM, Eisen C, Stenzinger A, et al. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med 2016;22:278-87. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017;32:185-203.e13. [Crossref] [PubMed]
- Wang A, Yue F, Li Y, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 2015;16:386-99. [Crossref] [PubMed]
- Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010;467:1114-7. [Crossref] [PubMed]
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014;14:275-91. [Crossref] [PubMed]
- Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov 2014;13:497-512. [Crossref] [PubMed]
- Sykes DB, Kfoury YS, Mercier FE, et al. Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell 2016;167:171-186.e15. [Crossref] [PubMed]
- Ordóñez-Morán P, Dafflon C, Imajo M, et al. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell 2015;28:815-29. [Crossref] [PubMed]
- Boyd AL, Reid JC, Salci KR, et al. Acute myeloid leukaemia disrupts endogenous myelo-erythropoiesis by compromising the adipocyte bone marrow niche. Nat Cell Biol 2017;19:1336-47. [Crossref] [PubMed]
- Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol Cancer Res 2009;7:1893-901. [Crossref] [PubMed]
- Croci L, Barili V, Chia D, et al. Local insulin-like growth factor I expression is essential for Purkinje neuron survival at birth. Cell Death Differ 2011;18:48-59. [Crossref] [PubMed]
- Hwang WL, Jiang JK, Yang SH, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol 2014;16:268-80. [Crossref] [PubMed]
- Varghese S, Whipple R, Martin SS, et al. Multipotent cancer stem cells derived from human malignant peritoneal mesothelioma promote tumorigenesis. PLoS One 2012;7:e52825. [Crossref] [PubMed]
- Huch M, Bonfanti P, Boj SF, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 2013;32:2708-21. [Crossref] [PubMed]
- Srivastava N, Kollipara RK, Singh DK, et al. Inhibition of cancer cell proliferation by PPARγ is mediated by a metabolic switch that increases reactive oxygen species levels. Cell Metab 2014;20:650-61. [Crossref] [PubMed]
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature 2013;501:328-37. [Crossref] [PubMed]
- Seifert L, Werba G, Tiwari S, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016;532:245-9. [Crossref] [PubMed]
- Boj SF, Hwang CI, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324-38. [Crossref] [PubMed]
- Kopp JL, von Figura G, Mayes E, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012;22:737-50. [Crossref] [PubMed]
- von Figura G, Fukuda A, Roy N, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 2014;16:255-67. [Crossref] [PubMed]
- Kreso A, O'Brien CA, van Galen P, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013;339:543-8. [Crossref] [PubMed]
- Min SY, Kady J, Nam M, et al. Human 'brite/beige' adipocytes develop from capillary networks, and their implantation improves metabolic homeostasis in mice. Nat Med 2016;22:312-8. [Crossref] [PubMed]
- Côté CD, Rasmussen BA, Duca FA, et al. Resveratrol activates duodenal Sirt1 to reverse insulin resistance in rats through a neuronal network. Nat Med 2015;21:498-505. [Crossref] [PubMed]
- Prost S, Relouzat F, Spentchian M, et al. Erosion of the chronic myeloid leukaemia stem cell pool by PPARγ agonists. Nature 2015;525:380-3. [Crossref] [PubMed]
- Stine RR, Shapira SN, Lim HW, et al. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol Metab 2016;5:57-65. [Crossref] [PubMed]
- Di Stefano B, Sardina JL, van Oevelen C, et al. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 2014;506:235-9. [Crossref] [PubMed]
- Marusyk A, Tabassum DP, Altrock PM, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 2014;514:54-8. [Crossref] [PubMed]
- Zammarchi F, Morelli M, Menicagli M, et al. KLF4 is a novel candidate tumor suppressor gene in pancreatic ductal carcinoma. Am J Pathol 2011;178:361-72. [Crossref] [PubMed]
- Galván JA, Zlobec I, Wartenberg M, et al. Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br J Cancer 2015;112:1944-50. [Crossref] [PubMed]
- Resnick MB, Konkin T, Routhier J, et al. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol 2005;18:511-8. [Crossref] [PubMed]
- Boumahdi S, Driessens G, Lapouge G, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 2014;511:246-50. [Crossref] [PubMed]
- Singh SK, Chen NM, Hessmann E, et al. Antithetical NFATc1-Sox2 and p53-miR200 signaling networks govern pancreatic cancer cell plasticity. EMBO J 2015;34:517-30. [Crossref] [PubMed]
- Krebs AM, Mitschke J, Lasierra Losada M, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 2017;19:518-29. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- Dowling RJ, Topisirovic I, Alain T, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 2010;328:1172-6. [Crossref] [PubMed]
- Wang S, Patsis C, Koromilas AE. Stat1 stimulates cap-independent mRNA translation to inhibit cell proliferation and promote survival in response to antitumor drugs. Proc Natl Acad Sci U S A 2015;112:E2149-55. [Crossref] [PubMed]
- Tsai SY, Rodriguez AA, Dastidar SG, et al. Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice. Cell Rep 2016;16:1903-14. [Crossref] [PubMed]
- Sun S, Chen S, Liu F, et al. Constitutive Activation of mTORC1 in Endothelial Cells Leads to the Development and Progression of Lymphangiosarcoma through VEGF Autocrine Signaling. Cancer Cell 2015;28:758-72. [Crossref] [PubMed]
- Rossi F, Gonzalez C. Synergism between altered cortical polarity and the PI3K/TOR pathway in the suppression of tumour growth. EMBO Rep 2012;13:157-62. [Crossref] [PubMed]
- Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 2014;13:140-56. [Crossref] [PubMed]
- Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer 2015;15:7-24. [Crossref] [PubMed]
- Morris JP 4th, Cano DA, Sekine S, et al. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest 2010;120:508-20. [Crossref] [PubMed]
- Birdsey GM, Shah AV, Dufton N, et al. The endothelial transcription factor ERG promotes vascular stability and growth through Wnt/β-catenin signaling. Dev Cell 2015;32:82-96. [Crossref] [PubMed]
- Su Y, Li J, Shi C, et al. N-cadherin functions as a growth suppressor in a model of K-ras-induced PanIN. Oncogene 2016;35:3335-41. [Crossref] [PubMed]
- Jeffery E, Church CD, Holtrup B, et al. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat Cell Biol 2015;17:376-85. [Crossref] [PubMed]
- Conde-Perez A, Gros G, Longvert C, et al. A caveolin-dependent and PI3K/AKT-independent role of PTEN in β-catenin transcriptional activity. Nat Commun 2015;6:8093. [Crossref] [PubMed]
- Laklai H, Miroshnikova YA, Pickup MW, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med 2016;22:497-505. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Diaferia GR, Balestrieri C, Prosperini E, et al. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J 2016;35:595-617. [Crossref] [PubMed]
- Elgogary A, Xu Q, Poore B, et al. Combination therapy with BPTES nanoparticles and metformin targets the metabolic heterogeneity of pancreatic cancer. Proc Natl Acad Sci U S A 2016;113:E5328-36. [Crossref] [PubMed]
- Zhao T, Jiang W, Wang X, et al. ESE3 Inhibits Pancreatic Cancer Metastasis by Upregulating E-Cadherin. Cancer Res 2017;77:874-85. [Crossref] [PubMed]
- Rajakumari S, Wu J, Ishibashi J, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab 2013;17:562-74. [Crossref] [PubMed]
- Kieslinger M, Folberth S, Dobreva G, et al. EBF2 regulates osteoblast-dependent differentiation of osteoclasts. Dev Cell 2005;9:757-67. [Crossref] [PubMed]
- Huang W, Wang G, Delaspre F, et al. Retinoic acid plays an evolutionarily conserved and biphasic role in pancreas development. Dev Biol 2014;394:83-93. [Crossref] [PubMed]
- Tenbaum SP, Ordóñez-Morán P, Puig I, et al. β-catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nat Med 2012;18:892-901. [Crossref] [PubMed]
- Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016;352:227-31. [Crossref] [PubMed]
- Baumgartner BK, Cash G, Hansen H, et al. Distinct requirements for beta-catenin in pancreatic epithelial growth and patterning. Dev Biol 2014;391:89-98. [Crossref] [PubMed]
- Sanchez-Gurmaches J, Hung CM, Sparks CA, et al. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab 2012;16:348-62. [Crossref] [PubMed]
- Eser S, Reiff N, Messer M, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013;23:406-20. [Crossref] [PubMed]
Cite this article as: Mathew H, Barb JJ, Bentzen S, Varghese S. EBF2 contributes to pancreatic cancer progenitor cell differentiation and tumor suppression. Ann Pancreat Cancer 2022;5:4.


